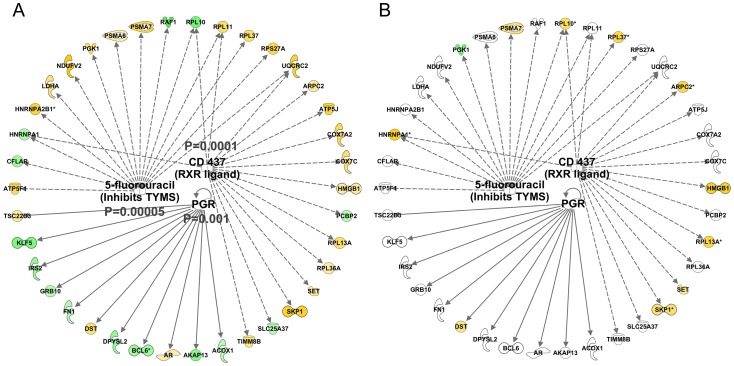
Figure 5. Quantitative SAM analysis using a continuum of age versus gene expression produces network hubs that are activated with human muscle age.
Using a total of 97 U133+2 Affymetrix gene-chips newly produced from two independent studies, the DRET study and the HERITAGE family study we produced a novel analysis that relied on the full age-range present in these data sets. A) We first found a set of genes that co-varied with age in the DRET study and then confirmed that 580 of these were also related to age in the HERITAGE study. Mitochondrial genes were not a feature of this linear age vs gene analysis. We then mapped the Affymetrix probe-sets to the IPA database and examined the up-stream analysis output. We found in IPA that the age-related dataset was consistent with the activation of the PGR (z-score = 2.6 and p-value = 0.001) and RXR (z-score = 2.0 and p-value = 0.0001) proteins and 5-fluorouracil agonism (Z-score = 2.2 and p-value = 0.0005). B) We noted that some members of these age-related networks were also associated with lean mass gains in humans. However about 50% of the common genes were positively associated with lean mass gain and age; and 50% were regulated in a discordant manner. Clearly some responses can be causal, some may be purely correlative and some may represent compensatory events.