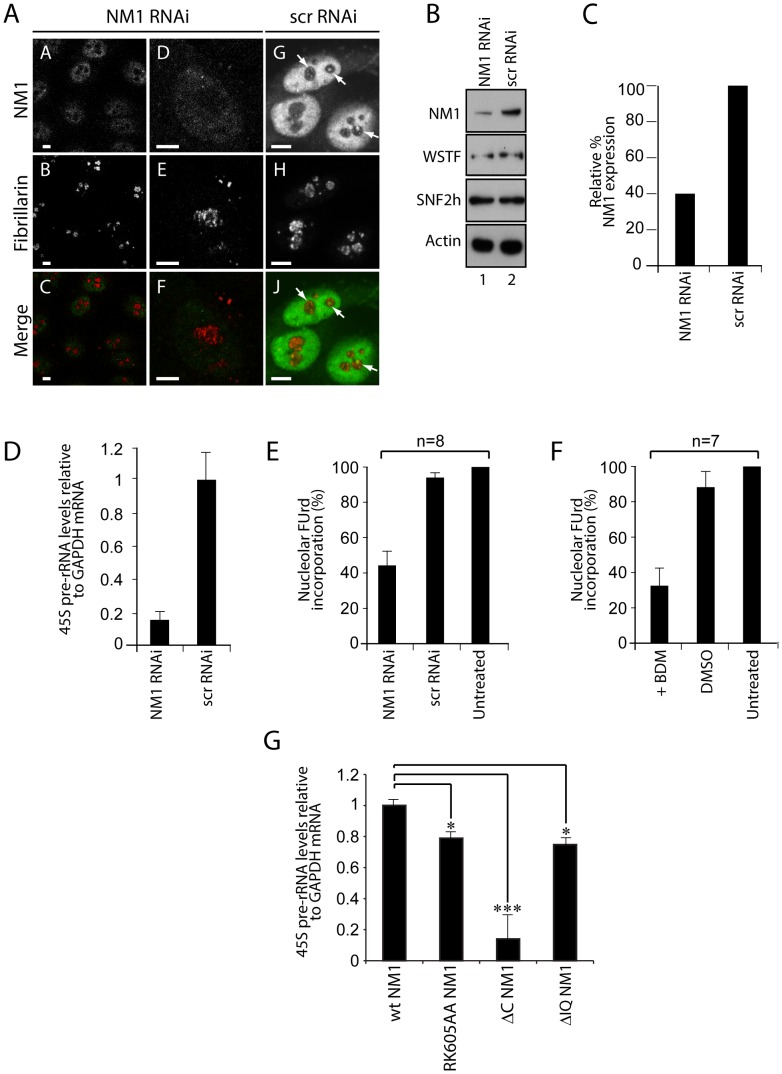
Figure 5. A functional NM1 is required for the activation of pol I transcription.
(A) RNAi-mediated NM1 gene knockdown in HeLa cells analyzed by double immunostaining and confocal microscopy with antibodies to NM1 and fibrillarin, after transfection of NM1-specific siRNA or control oligos (scrRNAi). Scale bars, 5 µm. (B) Steady state expression levels for NM1, WSTF, SNF2h and actin monitored on immunoblots of total lysates prepared from HeLa cells subjected to NM1 gene knockdown (NM1 RNAi) or from cells subjected to scrRNAi. (C) Semiquantitative densitometric quantification of NM1 steady state protein expression relative to actin. (D) qRT-PCR analysis of 45S pre-rRNA performed on total RNA prepared from HeLa cells subjected to NM1 gene knockdown (NM1 RNAi) or from cells subjected to control siRNA oligonucleotides (scrRNAi). The 45S pre-rRNA levels are relative to GAPDH mRNA. (E–F) FUrd incorporation assays on living HeLa cells subjected to (E) NM1 gene knockdown by RNAi, or (F) treated with BDM. Transcription was monitored by short FUrd pulses to follow incorporation into nascent nucleolar transcripts. Quantification of the FUrd foci after immunostaining and confocal microscopy was performed by measurements on randomly selected nucleolar regions in the images. The signal was quantified using ImageJ software. The average of the mean grey values in control cells was determined, and defined as hundred percent of signal. The average of the mean grey values measured after treatment was expressed proportionally. n = number of cells in each experiment. Error bars represent standard deviations. (G) rRNA synthesis in HEK293T cells stably expressing V5-wtNM1, V5-RK605AA, V5-ΔC or V5-ΔIQ NM1 mutants. For the analysis, relative 45S pre-rRNA levels were monitored from total RNA preparations by RT–qPCR using GAPDH mRNA as internal control. Error bars represent the standard deviation of three independent experiments. Significances [p RK605AA NM1 = 0.019 (*), p ΔC NM1 = 0.0006 (***), p ΔIQ NM1 = 0.05 (*)] were obtained by Student's T-test, two-sample, equal variance.