Figure 5.
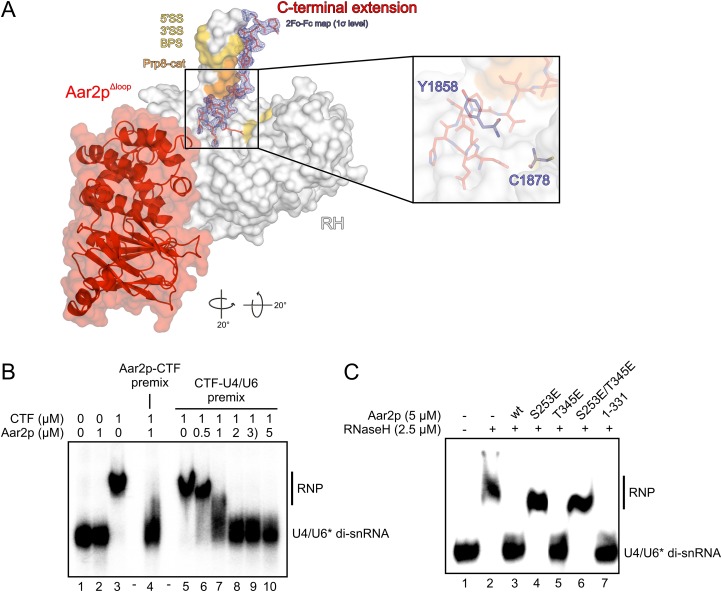
Aar2p interferes with U4/U6 di-snRNA binding to CTF. (A) Binding of the Aar2p C terminus (in sticks) along the thumb of the RH domain (in surface representation). (Gold) Prp8p residues that, upon mutation, suppress defects in either splice site or the branch point sequence of a pre-mRNA (Umen and Guthrie 1995, 1996; Collins and Guthrie 1999; Siatecka et al. 1999; Query and Konarska 2004); (orange) Prp8-cat residues (Grainger and Beggs 2005), which, upon mutation, suppress u4-cs1 phenotypes (Kuhn et al. 1999; Kuhn and Brow 2000). The final 2Fo − Fc electron density contoured at the 1σ level around the Aar2p C terminus is shown as a blue mesh. Rotation symbols indicate the views relative to Figure 1B. (Inset) Position of Prp8p residues Y1858 and C1878 under the Aar2p C terminus, which could be UV-cross-linked in vitro to U4 snRNA and U6 snRNA, respectively (Mozaffari-Jovin et al. 2012). (B) EMSA monitoring binding of U4/U6 di-snRNA in the absence and presence of Aar2p. Protein names and concentrations are given at the top of the gel. (Premix) Preincubation of the indicated components; (U6*) [32P]-labeled U6 snRNA. (C) EMSA monitoring the effects of phospho-mimetic mutations (S253E, T345E, and S253E/T345E) and of a C-terminal deletion in Aar2p (Aar2p1–331) on its ability to compete with U4/U6 di-snRNA for RH.