Figure 2.
High-Resolution Mapping of the Causative Locus
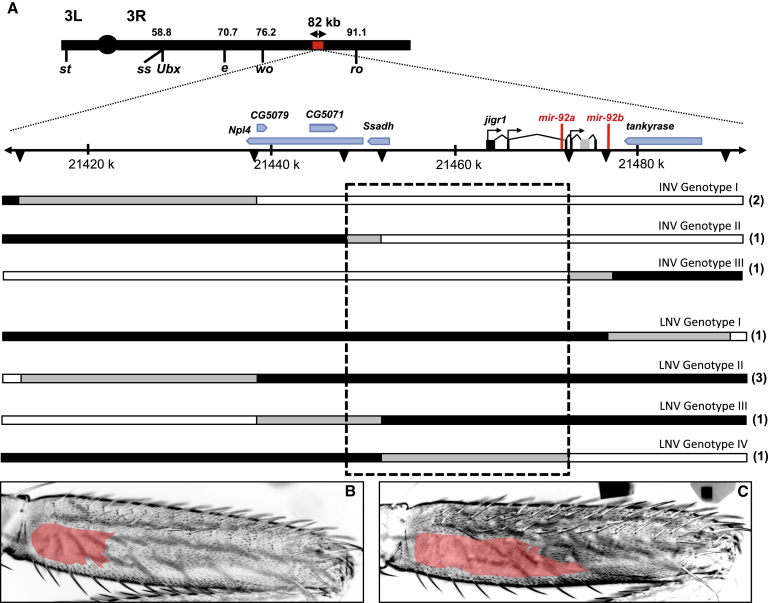
(A) The topmost black bar represents chromosome 3, with the two arms (3L and 3R) indicated either side of the centromere (circle). The position of Ubx and selected QTL markers are shown below the bar, with their positions in cM indicated above. The section of chromosome 3R highlighted by the red bar represents the 82.2 kb evolved region identified by the first mapping experiment (Figure S2C), which is shown expanded below, and between the dotted diagonal lines, with the scale given in kb. The bars below the scale indicate the genotypes of selected recombinants with breakpoints in the 82.2 kb region (note that all flies also carried a nonrecombinant chromosome from strain e, wo, ro that is not shown). Positions of molecular markers (Table S2) are indicated by black triangles. The number of individual flies representing each of the selected recombinant genotypes illustrated is given in parentheses to the right. Chromosomal regions from strains e, wo, ro (large naked valley parental line) and Oregon-R (small naked valley parental line) are indicated in black and white, respectively. Chromosomal regions in gray indicate DNA where the parental strain identity was not determined. The dashed black box indicates the 25 kb region that underlies naked valley variation. INV and LNV indicate intermediate and large naked valley phenotypes, respectively.
(B and C) Representative examples of T2 posterior femurs from recombinant flies with either an INV (B) or LNV (C).