Figure 4.
Differential Expression of mir-92a Underlies Naked Valley Variation through Repression of sha
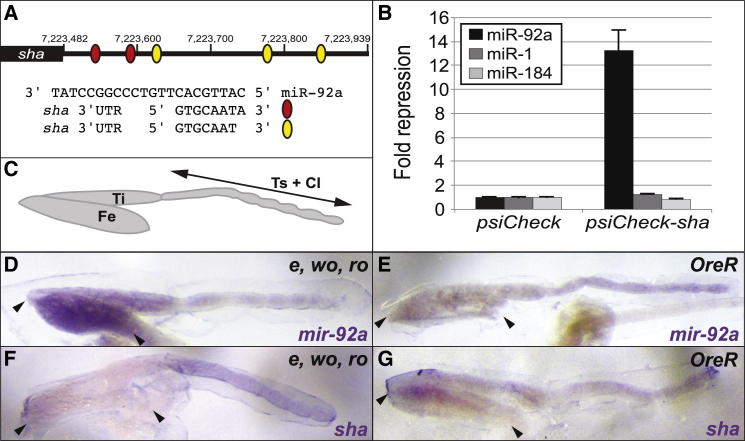
(A) The sha 3′ UTR contains five highly conserved, canonical seed-match sites for miR-92a (see also Figure S4). The black rectangle represents the sha coding region and the black line the 3′ UTR. Numbering is with respect to the base-pair position on chromosome 2R. Red and yellow ovals represent predicted seed-match sites for miR-92a consensus sequences shown aligned with the mature miR-92a sequence.
(B) Luciferase sensor assays in S2 cells show that the sha 3′ UTR confers >13-fold repression in response to ectopic miR-92a but is unaffected by control miR-1 and miR-184. Error bars show the SD from four independent transfections.
(C) Representation of the T2 pupal leg showing the femur (Fe), tibia (Ti), tarsa (Ts), and claws (Cl).
(D–G) Expression of pri-mir-92a (D and E) and sha (F and G), in the pupal T2 legs of strains e, wo, ro and Oregon-R at 24 hr APF. Arrowheads indicate the femur in each picture.