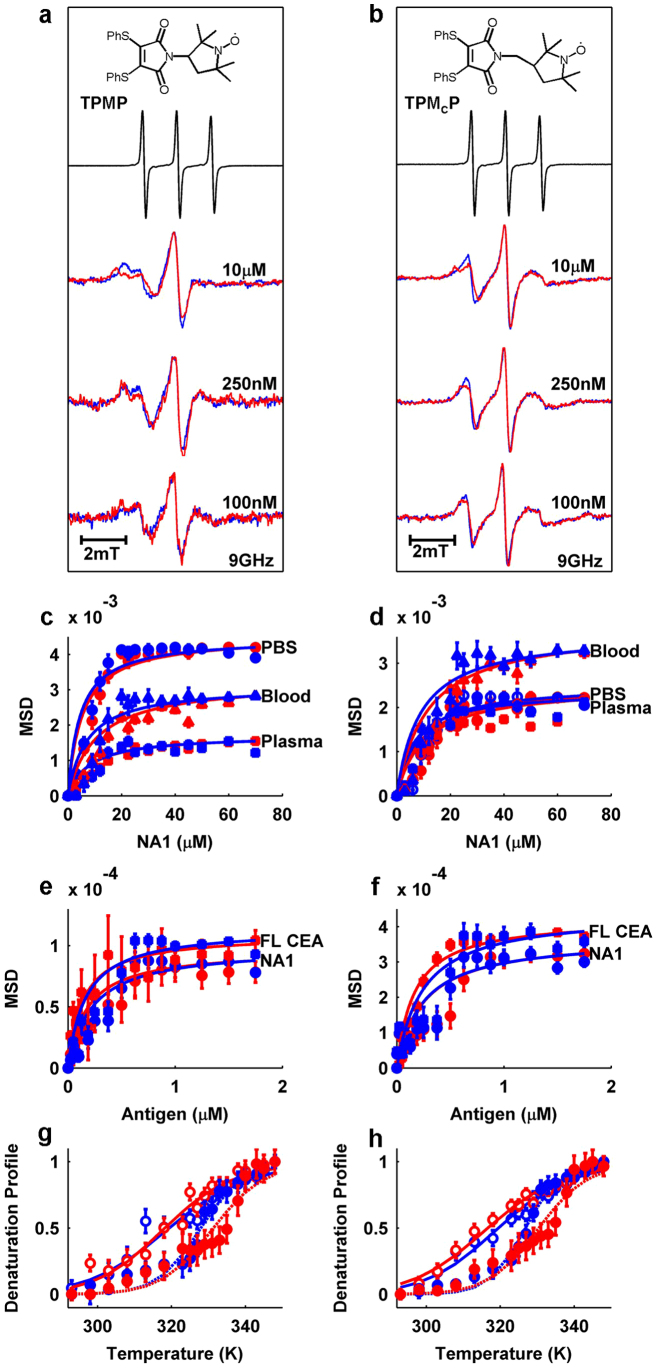
Figure 4. EPR-based monitoring of antibody-antigen interactions.
(a) From top to bottom: TPMP molecule, free TPMP normalized EPR spectrum (black), MP-sscFv (blue, 10 μM) and NA1-bound state (red, 70 μM NA1) EPR spectra in human blood, MP-sscFv (blue, 250 nM) and NA1-bound state (red, 1.75 μM NA1) EPR spectra in human blood, MP-sscFv (blue, 100 nM) and NA1-bound state (red, 400 nM NA1) EPR spectra in human blood. (b) Same as (a) but for MCP-sscFv. (c) 10 μM MP-sscFv and (d) MCP-sscFv binding curves. NA1 concentrations are 0, 3, 6, 9, 12, 15, 20, 22.5, 25, 30, 35, 40, 45, 50, 60 and 70 μM. Red circles (PBS), squares (human plasma), triangles (whole human blood) represent the respective raw data points and red lines represent best fits for raw data. Blue shapes and lines represent data points for simulations and fits of simulated data, respectively. (e) 250 nM binding curves for MP-sscFv and (f) MCP-sscFv with NA1 (circles) and full length CEA (squares), respectively. Fits are as described for c and d. NA1 and full length CEA concentrations are 0, 25, 50, 75, 100, 125, 187.5, 250, 375, 500, 625, 750, 875, 1000, 1250, 1500 and 1750 nM (calculated and simulated Kd values are listed in Supplementary Table S1). (g) MP-sscFv (blue, open circles) and MP-sscFv-SM3E (red, open circles) thermal stability shift assay with (closed circles) and without NA1 (70 μM) present. Tm values were fit using Supplementary Equation S2. (h) Same as g but for MCP-sscFv and MCP-sscFv-SM3E. Tm shifts are mentioned in the main text and all other values and enthalpies of unfolding are listed in Supplementary Table S2. Raw data represent the ratio of the low field and central nitroxide peaks. Error bars shown represent the 95% confidence boundaries as calculated from the data fitting process.