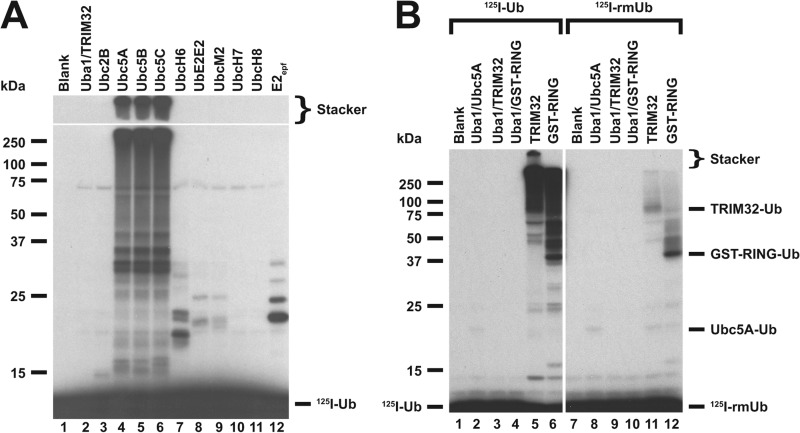
FIGURE 1.
E2 screen for TRIM32-catalyzed 125I-ubiquitin conjugation. A, autoradiogram of SDS-PAGE-resolved assays of TRIM32-catalyzed 125I-ubiquitin chain formation. Assays were conducted as described under “Materials and Methods” in the presence of 44 nm Uba1, 50 nm of the indicated E2, and 800 nm TRIM32. B, autoradiogram of SDS-PAGE resolved assays conducted identically to panel A but in the presence of 1.6 μm TRIM32 or 27 μm GST-RING protein and 460 nm Ubc5A as indicated. Incubations additionally contained 5 μm wild type (left subpanel) or reductively methylated (rmUB, right subpanel) 125I-ubiquitin. The left and right panels were exposed to normalize to account for the difference in specific radioactivity of the polypeptides. For panels A and B, migration positions for relative molecular weight standards are shown to the left. The position of the 5% stacker gel (Stacker), monoubiquitinated TRIM32 (Mwcalc = 80 kDa), monoubiquitinated GST-RING (Mwcalc = 44 kDa), monoubiquitinated Ubc5A (Mwcalc = 25 kDa), and free 125I-ubiquitin are shown to the right. Deviation of the apparent relative molecular weight of the monoubiquitinated species from their calculated molecular masses (Mwcalc) reflects non-ideality of ubiquitin due to its partial unfolding in SDS.