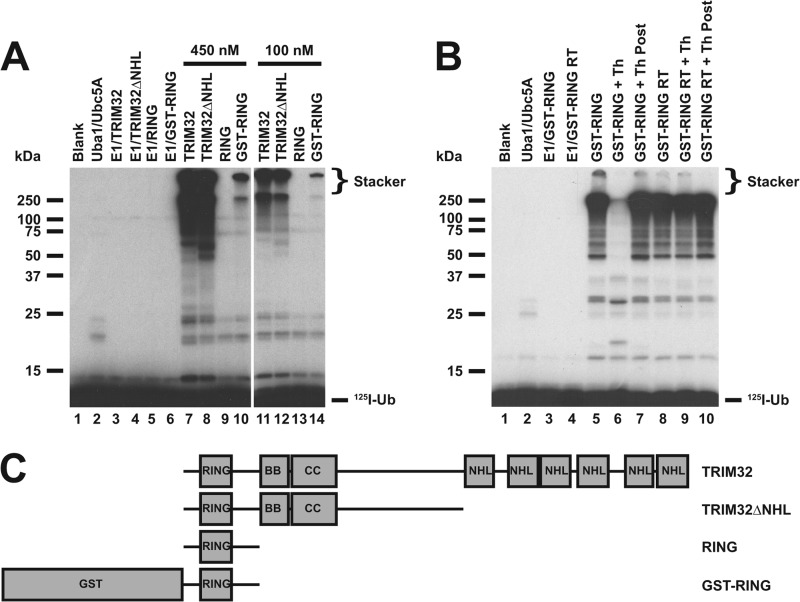
FIGURE 3.
Polyubiquitin chain formation requires multimerization. A, incubations similar to those of Fig. 1 were conducted with TRIM32 (lanes 7 and 11), TRIM32ΔNHL (lanes 8 and 12), RING domain alone formed by processing with 90 IU/ml of thrombin for 30 min (lanes 9 and 13), or a GST-RING fusion (lanes 10 and 14). Reactions contained 48 nm Uba1 and either 450 (left subpanel) or 100 nm (right subpanel) of the indicated E3 protein in the absence (lanes 3–6) or presence (lanes 7–14) of 280 nm Ubc5A. B, thrombin-dependent processing of GST from the RING moiety is responsible for abrogation of chain formation. Conjugation reactions contained 34 nm Uba1 and 18 μm GST-RING protein in the absence (lanes 3 and 4) or presence of 600 nm Ubc5A (lanes 4–10). The GST-RING or GST-RING ruined thrombin in which the thrombin cleavage site had been mutated to obviate processing were preincubated in the absence (lanes 5 and 8) or presence (lanes 6 and 9) of 90 IU/ml of thrombin for 30 min before addition to the conjugation assay. Alternatively, thrombin treatment was conducted after quenching the conjugation reaction (lanes 7 and 10). For both panels, migration positions for relative molecular weight standards are shown to the left. The position of the 5% stacking gel (Stacker), free 125I-ubiquitin, and mono-125I-ubiquitinated RING are shown to the right. C, schematic representation of constructs used in panels A and B.