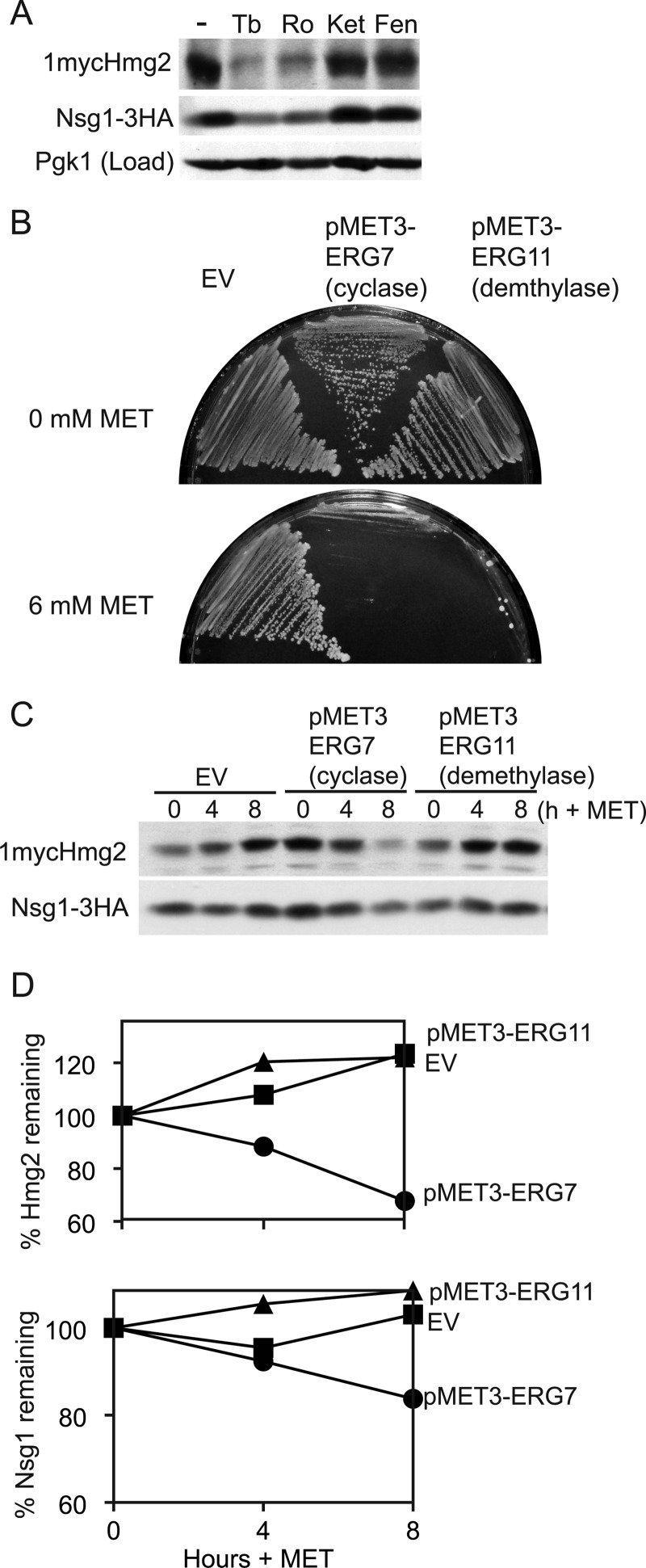
FIGURE 2.
Lanosterol synthesis promotes Hmg2-Nsg1 stability. A, inhibition of lanosterol synthesis destabilized Hmg2 and Nsg1. Wild-type cells grown to log phase in minimal medium were treated with inhibitors for 4 h at 30 °C, lysed with SUME, and subjected to SDS-PAGE followed by immunoblotting for 1mycHmg2, Nsg1–3HA, and Pgk1 (load control). The following concentrations of inhibitors were used: Tb, 10 μg/ml; Ro48–8071 (Ro), 40 μg/ml; ketoconazole (Ket), 10 μg/ml; fenpropimorph (Fen), 10 μg/ml. The experiment was performed at least three times. A typical result is shown. B, methionine repression of ERG7 (lanosterol cyclase) or ERG11 (lanosterol demethylase) genes efficiently kills yeast cells. Strains prototrophic for methionine production were transformed with URA3 empty vector (EV) or MET3-ERG7 or MET3-ERG11 plasmids, and growth was assayed in the absence (top) or presence (bottom) of 6 mm methionine. ERG7 and ERG11 are essential genes, and their growth is accordingly prohibited by the addition of methionine, demonstrating the effectiveness of the repression system. C, the same strains as in B were grown to log phase in minimal medium lacking methionine, and then 6 mm methionine was added to half of each culture. Samples were taken at the indicated time points, lysed and subjected to SDS-PAGE followed by immunoblotting. D, quantitations of protein levels in C were performed with ImageJ. Raw integrated density values were used to calculate the percentage remaining as for Fig. 1. Similar experiments were performed at least three times. Protein levels in strains carrying empty vector (squares), pMET3-ERG7 (circles), and pMET3-ERG11 (triangles) are plotted.