Table 1.
Structure of compounds, relative maximal ERK activity and hydrogen bonding energy between compounds and the homology model of the FFA1 receptor or GPR120
| Compound | Structure | Relative maximum ERK response | H-bonding energy (arbitrary units) | ||
|---|---|---|---|---|---|
| FFA1 | GPR120 | FFA1 | GPR120 | ||
| α-LA | 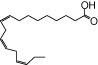 |
1.00 | 1.00* | −5.73 | −3.03* |
| AMGEN-1 | 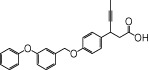 |
1.25 | 0.13 | −7.48 | −3.15 |
| GW9508 | 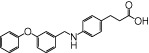 |
0.80 | 0.28 | −4.79 | −2.83 |
| MEDICA16 | 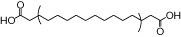 |
0.90 | 0.21* | −5.91 | −1.73* |
| NCG20 | 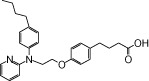 |
0.93 | 1.38* | −4.85 | −4.04* |
| NCG21 | 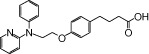 |
0.67 | 1.54* | −5.09 | −5.92* |
| NCG22 | 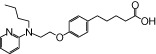 |
0.64 | 0.93* | −5.17 | −5.07* |
| NCG23 | 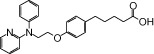 |
1.07 | 1.20* | −6.58 | −4.10* |
| NCG25 | 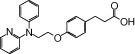 |
1.26 | 1.25* | −7.16 | −4.73* |
| NCG26 | 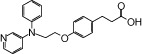 |
1.10 | 1.02* | −5.87 | −4.54* |
| NCG27 | 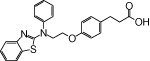 |
0.61 | 0.55* | −3.50 | −2.23* |
| NCG28 | 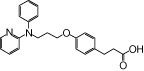 |
1.20 | 0.80* | −5.69 | −3.66* |
| NCG29 | 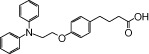 |
0.17 | 1.46* | −4.19 | −3.56* |
| NCG30 | 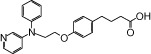 |
0.36 | 1.45* | −3.46 | −5.57* |
| NCG31 | 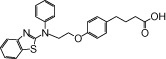 |
0.13 | 1.40* | −3.22 | −4.06* |
| NCG32 | 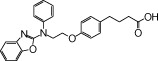 |
0.87 | 1.03* | −5.34 | −3.86* |
| NCG33 | 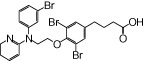 |
0.04 | 0.39* | −3.80 | −2.88* |
| NCG34 | 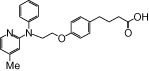 |
1.17 | 1.37* | −5.78 | −5.36* |
| NCG35 | 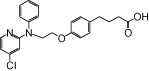 |
1.30 | 1.48* | −7.53 | −4.75* |
| NCG38 | 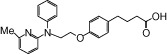 |
1.03 | 1.24* | −6.87 | −4.87* |
| NCG39 | 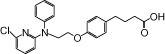 |
0.42 | 1.23* | −4.09 | −4.75* |
| NCG40 | 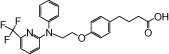 |
1.03 | 0.62* | −5.42 | −2.73* |
| NCG41 | 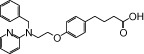 |
1.61 | 0.96* | −7.95 | −2.61* |
| NCG42 | 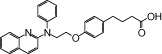 |
0.79 | 0.54* | −3.46 | −2.92* |
| NCG44 | 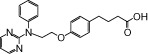 |
0.32 | 0.64* | −4.30 | −3.10* |
| NCG45 | 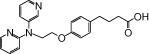 |
0.06 | 0.76* | −3.33 | −3.39* |
| NCG46 | 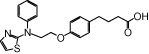 |
0.53 | 1.20* | −5.21 | −5.41* |
| NCG48 | 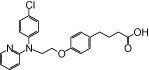 |
1.04 | 0.92* | −6.15 | −3.98* |
| NCG49 | 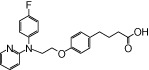 |
0.89 | 0.75* | −5.23 | −2.61* |
| NCG50 | 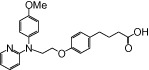 |
1.02 | 1.14* | −3.96 | −4.93* |
| NCG51 | 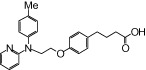 |
0.95 | 1.32* | −5.75 | −4.77* |
| NCG52 | 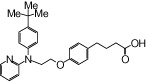 |
0.90 | 0.29* | −4.29 | −2.70* |
| NCG53 | 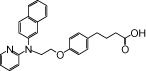 |
1.47 | 1.27* | −5.82 | −4.56* |
| NCG57 | 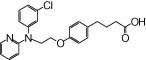 |
1.21 | 1.09* | −6.98 | −4.11* |
| NCG74 | 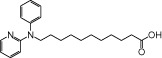 |
1.64 | 0.66 | −7.05 | −1.67 |
| NCG75 | 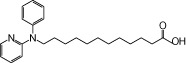 |
1.63 | 0.27 | −6.77 | −2.19 |
| NCG85 | 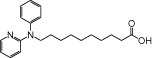 |
1.45 | 0.86 | −6.85 | −3.52 |
| NCG86 | 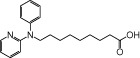 |
0.62 | 0.74 | −3.37 | −2.84 |
| NCG87 | 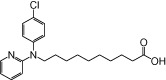 |
0.99 | 0.10 | −5.13 | −1.11 |
| NCG88 | 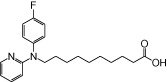 |
0.91 | 0.20 | −6.89 | −3.42 |
| NCG89 | 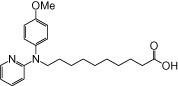 |
0.86 | 0.11 | −6.87 | −0.76 |
| NCG97 | 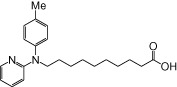 |
1.20 | 0.22 | −6.57 | −1.21 |
| NCG98 | 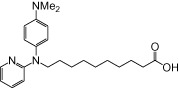 |
0.89 | 0.19 | −5.24 | −1.65 |
| NCG99 | 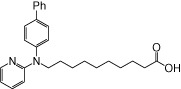 |
0.87 | 0.18 | −5.82 | −2.34 |
| NCG100 | 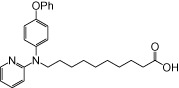 |
0.18 | 0.11 | −2.22 | −3.08 |
| NCG101 | 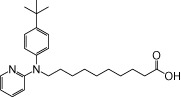 |
0.70 | 0.04 | −4.36 | −1.50 |
| NCG102 | 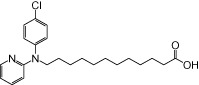 |
0.80 | 0.05 | −5.36 | −1.27 |
| NCG103 | 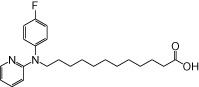 |
0.66 | 0.12 | −3.24 | −2.54 |
| NCG104 | 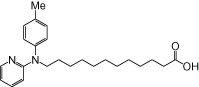 |
0.63 | 0.02 | −5.93 | −1.00 |
| NCG105 | 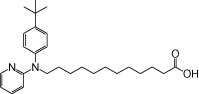 |
0.38 | 0.04 | −3.19 | −1.45 |
| NCG106 | 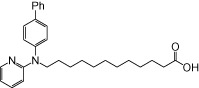 |
0.66 | 0.07 | −4.24 | −0.18 |
The relative maximal ERK response was calculated as the relative value of the agonist response elicited by each compound (100 µM), with respect to the response evoked by 100 µM α-LA in FFA1 receptor- or GPR120-expressing cells.
Values from Sun et al. (2010).
