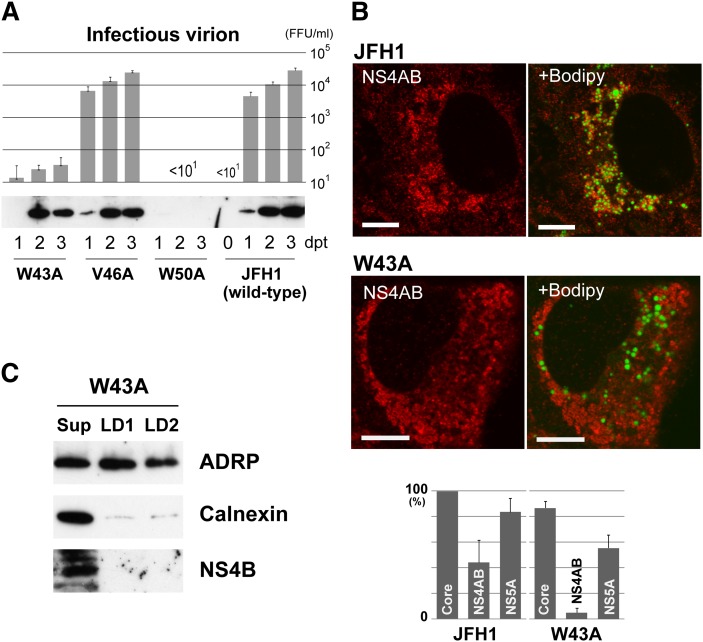
Fig. 5.
JFH1 mutants harboring single alanine substitutions for hydrophobic residues of NS4B are defective for replication. (A) Effect on virus replication of alanine substitutions in NS4B region of JFH1. (Top) Titration of infectious virions released from transfectants of JFH1 or mutant RNAs. Culture supernatant of each transfectant was assayed for infectivity at the indicated days post transfection (dpt). FFU, focus forming unit. Mean values from three or four independent experiments are shown; error bars denote SD. (Bottom) Expression of NS4B in the transfectants of JFH1 or mutant RNAs. (B) Localization of HCV antigens and the changes caused by W43A mutation. (Top) Change in localization of NS4AB (NS4B and/or NS4A). Transfectants with JFH1 or W43A RNA were stained with anti-NS4AB antibody, followed by staining with Bodipy 493/503. Scale bars, 10 μm. (Bottom) Comparison of LD association for HCV antigens between W43A- and JFH1-RNA-transfected cells. The percentages of positive cells for LD association of each antigen are shown. When association was seen for at least six LDs in a section, the transfectant was counted as positive. Mean values from three independent experiments are shown; error bars denote SD. (C) Localization of NS4B to LD preparations obtained from W43A-RNA-transfected cells. LD fractions were prepared and analyzed as in Fig. 2, right.