Abstract
Telomeres, the ends of linear eukaryotic chromosomes, shorten due to incomplete DNA replication and nucleolytic degradation. Cells counteract this shortening by employing a specialized reverse transcriptase called telomerase, which uses deoxyribonucleoside triphosphates (dNTPs) to extend telomeres. Intracellular dNTP levels are tightly regulated, and perturbation of these levels is known to affect DNA synthesis. We examined whether altering the levels of the dNTP pools or changing the relative ratios of the four dNTPs in Saccharomyces cerevisiae would affect the length of the telomeres. Lowering dNTP levels leads to a modest shortening of telomeres, while increasing dNTP pools has no significant effect on telomere length. Strikingly, altering the ratio of the four dNTPs dramatically affects telomere length homeostasis, both positively and negatively. Specifically, we find that intracellular deoxyguanosine triphosphate (dGTP) levels positively correlate with both telomere length and telomerase nucleotide addition processivity in vivo. Our findings are consistent with in vitro data showing dGTP-dependent stimulation of telomerase activity in multiple organisms and suggest that telomerase activity is modulated in vivo by dGTP levels.
Keywords: telomeres, telomere length homeostasis, dNTPs, telomerase, processivity
ALL eukaryotes, as well as some prokaryotes with linear chromosomes, contain repetitive sequences called telomeres at the ends of their DNA. Telomeric DNA is bound by proteins that protect chromosome ends from being recognized as genotoxic DNA double-strand breaks in need of repair (Jain and Cooper 2010). However, telomeres shorten due to incomplete DNA replication and nucleolytic degradation. Left unchecked, this telomere erosion eventually results in very short, unprotected telomeres, leading to cell-cycle arrest and replicative senescence (Lundblad and Szostak 1989; Harley et al. 1990; Yu et al. 1990).
Telomere shortening is counteracted by a specialized reverse transcriptase called telomerase (Greider and Blackburn 1985), whose core consists of a protein catalytic subunit and an RNA moiety—hTERT and hTR, respectively, in humans (Feng et al. 1995; Nakamura et al. 1997), and Est2 and TLC1, respectively, in the budding yeast Saccharomyces cerevisiae (Singer and Gottschling 1994; Lingner et al. 1997). Telomerase extends telomeres by repeated reverse transcription of a short sequence to the 3′ ends of telomeres, using the RNA subunit as a template (Greider and Blackburn 1989; Yu et al. 1990; Singer and Gottschling 1994). Although the sequence of the telomeric repeats differs between species, a common feature is that they are all G-rich. In vertebrates, the repeat sequence is TTAGGG (Meyne et al. 1989), while in S. cerevisiae, the telomeric repeats have a consensus sequence of (TG)0-6TGGGTGTG(G)0-1 (Forstemann and Lingner 2001).
Deoxyribonucleoside triphosphates (dNTPs) are the building blocks of DNA, and their production needs to be tightly regulated as imbalances in dNTP pools can be mutagenic (Reichard 1988). In S. cerevisiae, the sole mode of dNTP production is through de novo dNTP synthesis, and the primary enzyme in this process is ribonucleotide reductase (RNR). RNR catalyzes the reduction of adenosine triphosphate (ADP), guanosine diphosphate (GDP), cytidine diphosphate (CDP), and uridine diphosphate (UDP) to their respective deoxy forms, which are then converted to deoxyadenosine triphosphate (dATP), deoxyguanosine triphosphate (dGTP), deoxycytidine triphosphate (dCTP), and deoxythymidine triphosphate (dTTP) (Reichard 1988). Since RNR is the rate-limiting step in dNTP production, it is tightly regulated to maintain proper dNTP levels. In S. cerevisiae, overall pool sizes are controlled by (1) allosteric regulation of RNR activity by dATP (Reichard 1988); (2) transcriptional regulation of the RNR genes by Crt1, Ixr1, as well as the MBF complex (Huang et al. 1998; de Bruin et al. 2006; Tsaponina et al. 2011); (3) direct inhibition of the large subunit of RNR, Rnr1, by Sml1 (Zhao et al. 1998; Chabes et al. 1999), and (4) relocalization of the small subunits of RNR, Rnr2, and Rnr4 to the nucleus by Wtm1 and Dif1 (Lee and Elledge 2006; Lee et al. 2008; Wu and Huang 2008). The regulation of the proteins Wtm1, Dif1, Crt1, and Sml1 is dependent on Mec1, the ATR ortholog in yeast, in a manner dependent on Dun1, a CHK2 ortholog (Huang et al. 1998; Zhao et al. 2001; Lee and Elledge 2006; Lee et al. 2008). In addition, an allosteric specificity site on RNR is important for maintaining the proper balance of the four individual dNTPs (Reichard 1988).
Previous studies have implicated dNTP pools in telomere length homeostasis. Both rnr1Δ and dun1Δ mutants were found to have short telomeres in a genome-wide screen that measured the telomere lengths of strains in the yeast gene deletion library (Gatbonton et al. 2006). Given that both Rnr1 and Dun1 are positive regulators of dNTP levels, the shortened telomeres of the rnr1Δ and dun1Δ mutants may be a product of reduced dNTPs. Indeed, the dNTP pool sizes in a dun1Δ strain are reduced twofold compared to wild type (Fasullo et al. 2010). Furthermore, cdc8 and cdc21 mutants, which express defective thymidylate kinase and thymidylate synthetase, respectively, involved in the production of dTTP, possess shortened telomeres (Adams and Holm 1996). All of these observations suggest a possible link between telomere length homeostasis and regulation of dNTP pools. In vitro studies of telomerase from S. cerevisiae (Peng et al. 2001; Bosoy and Lue 2004), Euplotes aediculatus (Hammond and Cech 1997, 1998), Tetrahymena thermophila (Hardy et al. 2001), and mammals (Morin 1989; Maine et al. 1999) also suggest that dGTP levels influence telomerase activity, but the in vivo relevance of these observations is unclear.
In this study, we systematically varied dNTP pool levels and observed that reducing the overall pool size does indeed result in shortened telomeres. Interestingly, increasing dNTP levels has no significant effect on telomere length homeostasis. Additionally, we show that Mec1 and Dun1 function in the same pathway to regulate dNTP pools and telomere length homeostasis. However, Mec1 also functions with Tel1, the yeast ATM ortholog, to regulate telomerase in a Dun1-independent manner. Furthermore, by using rnr1 mutants that perturb the balance of the four dNTPs, we find dramatic effects on telomere length homeostasis, both positive and negative. In particular, intracellular dGTP levels positively correlate with telomere length and telomerase nucleotide addition processivity in vivo, consistent with in vitro data showing that dGTP levels can stimulate telomerase activity in yeast (Peng et al. 2001; Bosoy and Lue 2004), ciliates (Hammond and Cech 1997, 1998; Hardy et al. 2001), and mammals (Morin 1989; Maine et al. 1999). Thus, our findings reveal an important link between telomere length homeostasis and dNTP pools and show that alterations in intracellular dGTP levels can modulate telomerase activity.
Materials and Methods
Yeast media and strains
Standard yeast media and growth conditions were used (Sherman 1991). Yeast strains used in this study are listed in Supporting Information, Table S1. All the rnr1 mutants were expressed from the endogenous RNR1 promoter. For reasons unknown, we were previously unable to generate viable haploid cells carrying the rnr1-Q288E allele (Kumar et al. 2010), but here we were able to generate the strain without any difficulties with the same methodology that we used to construct the other rnr1 mutants.
Telomere PCR and sequencing
Yeast genomic DNA was isolated using a Yeast DNA Extraction Kit (Thermo Scientific) or Wizard Genomic DNA Purification Kit (Promega). Y′ telomeres and telomere VI-R were amplified by PCR as previously described (Pardo et al. 2006; Chang et al. 2007). Telomere VI-R PCR products were cloned using a PCR Cloning Kit (Qiagen). Sequencing was performed by GENEWIZ and BaseClear, and the resulting sequence data were analyzed using Sequencher software (Gene Codes).
Telomere length measurements
Telomeres, whether in a single cell or in a population of cells, exhibit a range of lengths, typically resulting in smears of ∼50–100 bp when analyzed after agarose gel electrophoresis. For each sample, the median telomere length (i.e., the most intense part of the smear) is determined. Then data from several isolates of each genotype are combined to determine the average of these median telomere lengths, as well as standard error of the mean values.
Determination of dNTP pools
dNTP levels were measured as previously described (Chabes et al. 2003).
Flow cytometry
Flow cytometry was performed as previously described (Sabouri et al. 2008).
Protein blotting
Protein extracts were prepared as previously described (Peter et al. 1993). For Rnr2, Rnr3, and Sml1 detection, affinity-purified rabbit polyclonal anti-Rnr2 (AS09 575), anti-Rnr3 (AS09 574), and anti-Sml1 (AS10 847) antibodies (Agrisera AB) were used at 1:500,000, 1:1000, and 1:5000 dilutions, respectively. For detection of both Rnr4 and α-tubulin, YL1/2 rat monoclonal antibody (Sigma) was used at a 1:2500 dilution (Tsaponina et al. 2011).
Results
Decreased dNTP pools lead to shortened telomeres
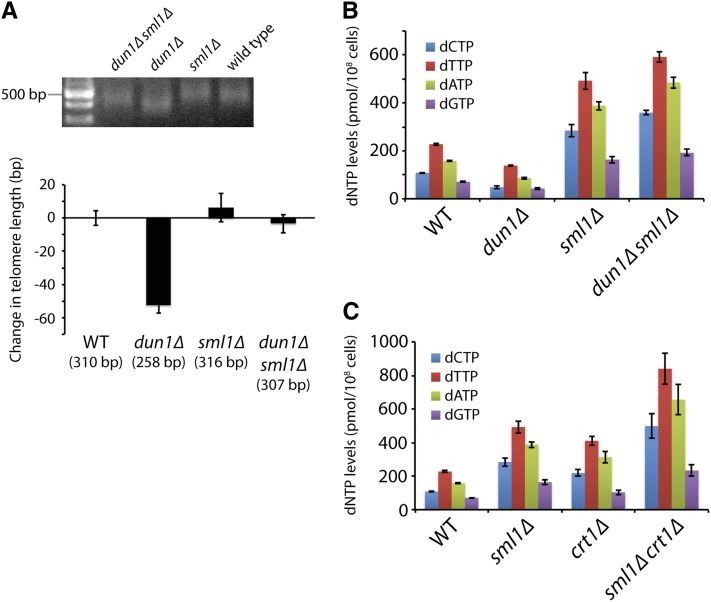
To determine whether altered dNTP pools affect telomere length homeostasis, we isolated meiotic products from the sporulation of a dun1Δ/DUN1sml1Δ/SML1 diploid and examined these haploid progeny for both telomere length and dNTP pool size. Consistent with previous observations (Gatbonton et al. 2006), the dun1Δ mutant shows slightly shorter telomeres compared to wild-type strains (∼50 bp shorter) (Figure 1A and Figure S1). This phenotype is rescued by additional deletion of SML1 (Figure 1A and Figure S1). Since dun1Δ strains have a 2-fold reduction in dNTP pools and dun1Δ sml1Δ double mutants show a 2.5-fold increase in dNTP pools similar to a sml1Δ mutant (Fasullo et al. 2010) (Figure 1B), the shortened telomeres in a dun1Δ mutant are likely caused by reduced dNTP pools. Interestingly, despite the increased dNTP pools in the sml1Δ and dun1Δ sml1Δ strains, there is no significant change in telomere length in either strain. Since deletion of CRT1 has previously been shown to increase dNTP pools 2-fold (Tang et al. 2009), we constructed sml1Δ crt1Δ strains hoping to further increase dNTP levels. Indeed, sml1Δ crt1Δ strains have total dNTP pools ∼4-fold above wild-type levels (Figure 1C), but the increase in telomere length is <20 bp (Figure S2A). Similarly, an rnr1-D57N mutant (Chabes et al. 2003) has dNTP pools almost 3-fold above wild-type levels and an increase in telomere length of ∼40 bp (Figure S2B), indicating that an increase in dNTP pools above wild-type levels does not dramatically affect telomere length homeostasis.
Figure 1.
Decreasing dNTP levels results in shorter telomeres. (A) Strains of the indicated genotypes, generated from the sporulation of a dun1Δ/DUN1 sml1Δ/SML1 diploid, were assayed for telomere length by Y′ telomere PCR after being passaged for at least 100 generations (a representative gel is shown). The change in telomere length, compared to wild-type telomere length, was quantified and plotted. Mean ± SE is shown for four independent isolates of each strain . Similar results were obtained by assaying telomere length by telomere I-L PCR (i.e., PCR amplification of the left telomere of chromosome I) and by denaturing in-gel hybridization (Figure S1). (B) Strains in A were assayed for dNTP levels. Four independent isogenic strains for each genotype were analyzed. Data are represented as mean ± SE. (C) Strains of the indicated genotypes were assayed for dNTP levels, as in B.
Elucidating the role of Mec1 at telomeres
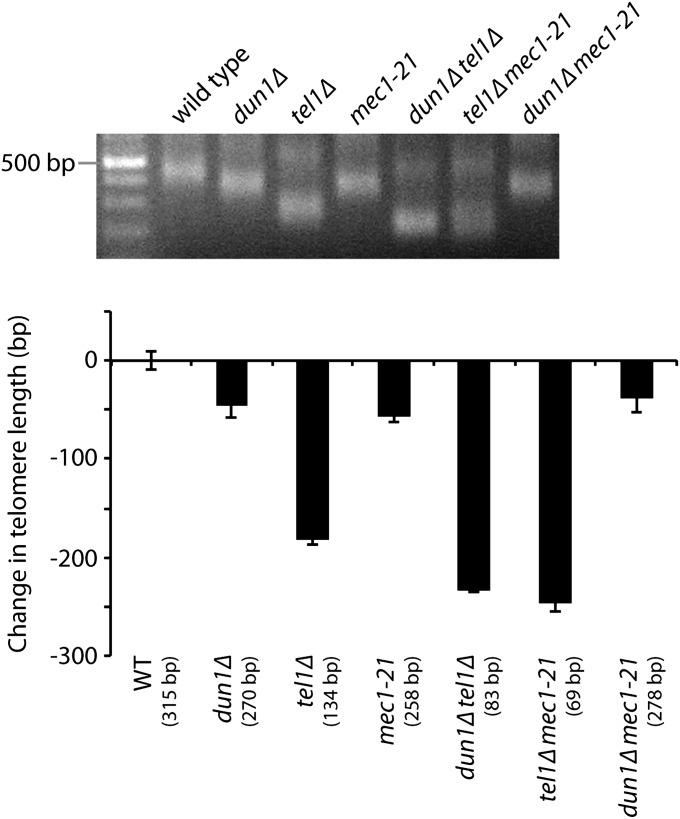
The phosphoinositide-3-kinase-related kinases Tel1 and Mec1, yeast orthologs of human ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related), respectively, are needed for proper regulation of telomerase (Ritchie et al. 1999; Arnerić and Lingner 2007). Yeast cells lacking Tel1 have very short telomeres (Greenwell et al. 1995), while a mec1-21 mutant has slightly short telomeres (Ritchie et al. 1999). Previous work has shown that mec1-21, dun1Δ, and dun1Δ mec1-21 mutants all have similar dNTP pool sizes (Fasullo et al. 2010), indicating that Mec1 also functions in a pathway with Dun1 to regulate dNTP pools. Therefore, we determined whether Mec1 and Dun1 also function in the same pathway to regulate telomere length homeostasis. We measured telomere length in a dun1Δ mec1-21 double mutant compared to the dun1Δ and mec1-21 single mutants, and all three strains showed the same telomere length (Figure 2). Thus, Mec1 and Dun1 function in the same pathway to regulate both dNTP pools and telomere length. In contrast, we find that Tel1 and Dun1 function in separate pathways to regulate telomere length since the effect of combining DUN1 and TEL1 deletions is additive: dun1Δ tel1Δ double mutants have shorter telomeres than either dun1Δ or tel1Δ single mutants (Figure 2).
Figure 2.
Mec1 and Dun1 function in the same pathway to regulate dNTP pools and telomere length homeostasis. Strains of the indicated genotypes, generated from the sporulation of a dun1Δ/DUN1 tel1Δ/TEL1 mec1-21/MEC1 diploid, were assayed for telomere length by Y′ telomere PCR after being passaged for ∼50 generations (a representative gel is shown). The change in telomere length, compared to wild-type telomere length, was quantified and plotted. Mean ± SE is shown for three independent isolates . Note that the triple mutant is not viable (data not shown).
We next examined these three genes for their role in senescence. It was previously shown that mec1-21 tel1Δ strains senesce due to a lack of telomerase-mediated telomere extension, a phenotype similar to that of telomerase-negative strains (Ritchie et al. 1999; Arnerić and Lingner 2007). Since MEC1 and DUN1 act in the same pathway for nucleotide pools and telomere length, we tested whether dun1Δ tel1Δ double mutants also senesce like mec1-21 tel1Δ strains. Interestingly, we find that dun1Δ tel1Δ cells do not senesce despite repeated subculturings (data not shown). These observations indicate that MEC1 and TEL1 function to prevent senescence in a pathway that is not dependent on Dun1.
Altering the ratio of the four dNTPs affects telomere length homeostasis
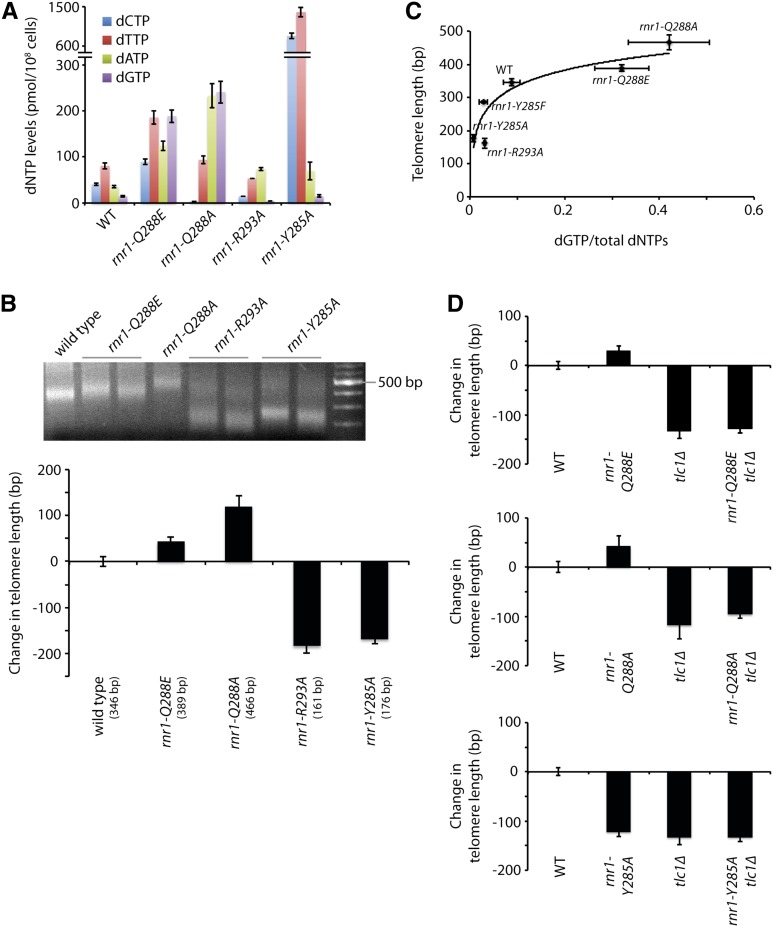
Having established that limiting dNTP pools leads to shorter telomeres, we explored whether perturbing the balance of the four dNTPs would also affect telomere length homeostasis. We have previously shown that the ratio of the four dNTPs can be severely imbalanced by making mutations in loop 2 of Rnr1 (Kumar et al. 2010). We focused on four rnr1 mutants—rnr1-Q288E, rnr1-Q288A, rnr1-R293A, and rnr1-Y285A—with imbalanced dNTP pools (Figure 3A). The rnr1 mutants were expressed from the endogenous RNR1 locus. It is important to note that, unlike the mutants previously mentioned (e.g., dun1Δ, sml1Δ, etc.) and all other mutants where dNTP pools have been measured, to the best of our knowledge, these rnr1 mutants are the only mutants known to cause an imbalance of the four dNTPs. We find that both the rnr1-Q288E and the rnr1-Q288A mutants show elongated telomeres compared to wild type while both the rnr1-R293A and the rnr1-Y285A mutants have dramatically shortened telomeres, almost 200 bp shorter than wild-type telomeres, which is much greater than the ∼50-bp decrease seen in the dun1Δ mutant (Figures 1A and 3B). These results indicate that disrupting the ratio of the four dNTPs can greatly affect telomere length homeostasis both positively and negatively. The increase in telomere lengths in the rnr1-Q288E and rnr1-Q288A mutants correlates with increases in total dNTP levels. However, the rnr1-Y285A mutant has very short telomeres despite having significantly increased total dNTP levels. Thus, the telomere length changes cannot be easily explained by changes in total dNTP levels (Figure S3A). Neither can growth rate provide an explanation since both the rnr1-Q288A and the rnr1-R293A mutants are delayed in S phase and grow very poorly (Kumar et al. 2010) but exhibit opposite effects on telomere length.
Figure 3.
Telomere length positively correlates with percentage of intracellular dGTP. (A) dNTP concentrations in a wild-type strain and the rnr1 mutants were measured. Mean ± SE is shown for each strain. Data for the wild type, rnr1-Q288A, rnr1-R293A, and rnr1-Y285A strains were previous reported (Kumar et al. 2010). (B) Strains of the indicated genotypes were assayed for telomere length by telomere VI-R PCR after being passaged for at least 100 generations (a representative gel is shown). The change in telomere length, compared to wild-type telomere length, was quantified and plotted. Mean ± SE is shown for at least three independent isolates. (C) Strains of the indicated genotype were plotted for telomere length vs. dGTP as a fraction of total dNTP levels. Each point indicates the mean for each of these values, and error bars indicate the standard error. (D) Strains of the indicated genotypes, generated from the sporulation of rnr1/RNR1 tlc1Δ/TLC1 diploids, were assayed for telomere length by Y′ telomere PCR after being passaged for ∼30 generations. The change in telomere length, compared to wild-type telomere length, was quantified and plotted. Mean ± SE is shown for three independent isolates.
Next, we compared the change in telomere length to the amount of each of the four dNTPs as a percentage of the total dNTP pool size. We found no correlation in length homeostasis in comparison to the dCTP, dTTP, and dATP pools in these strains (Figure S3A). On the other hand, we found a solid association between telomere length and dGTP pools: both the rnr1-R293A and the rnr1-Y285A mutants, which have short telomeres, show reduced dGTP while the two mutants with long telomeres, rnr1-Q288E and rnr1-Q288A, have increased dGTP as a percentage of the total dNTP levels (Figure 3C). These results indicate that telomere length is correlated with the percentage of intracellular dGTP more so than the absolute size of the dNTP pools. This correlation remained true when we further analyzed the dNTP pools and telomere length of a fifth rnr1 mutant, rnr1-Y285F (Figure 3C and Figure S3A). Consistent with this idea, the mutants analyzed in Figure 1 do not have altered percentages of dGTP (Figure S3B), resulting in much milder effects on telomere length homeostasis. Interestingly, if we exclude one of the five mutants (i.e., rnr1-R293A), it appears that telomere length is inversely correlated with dCTP and dTTP levels and positively correlated with dATP levels. However, in vitro data (see below) do not support this view.
To determine whether telomere length changes in the rnr1-Q288E, rnr1-Q288A, rnr1-R293A, and rnr1-Y285A mutants are dependent on telomerase, we made double mutants of the rnr1 point mutants with tlc1Δ by isolating haploid meiotic progeny after the sporulation of rnr1/RNR1tlc1Δ/TLC1 diploids. We were unable to carefully measure the telomere length of rnr1-R293A tlc1Δ double mutants as isolates of this strain senesced before we were able to extract DNA. In the one double-mutant strain that grew (of a possible 15), the telomeres were extremely short as measured by sequencing (see below). Thus, this synergistic interaction suggests that an additional factor(s) contributes to the shortened telomeres of the rnr1-293A mutant. In contrast, the other double mutants, rnr1-Q288E tlc1Δ, rnr1-Q288A tlc1Δ, and rnr1-Y285A tlc1Δ, all exhibit telomere lengths that are similar to tlc1Δ single mutants (Figure 3D), indicating that the telomere length changes associated with the rnr1-Q288E, rnr1-Q288A, and rnr1-Y285A mutants are all telomerase-dependent.
Altering dGTP affects telomerase nucleotide addition processivity
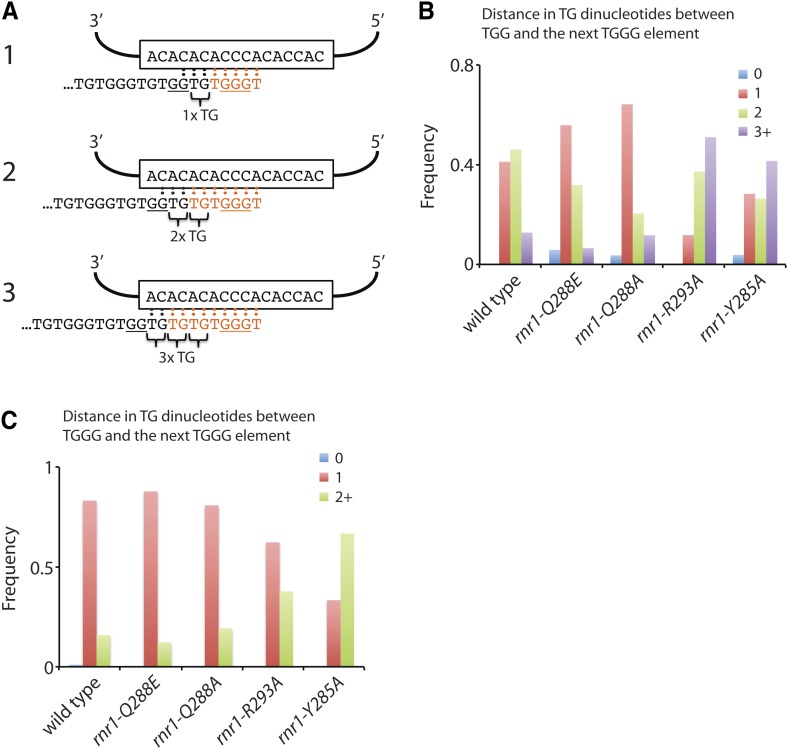
Since the telomere length changes were telomerase-dependent in the three rnr1 mutants that we could test, we examined the effect of imbalanced dNTP pools on the efficiency with which telomerase adds dNTPs to the 3′ terminus of telomeres (i.e., the nucleotide addition processivity of telomerase). Although the S. cerevisiae telomerase RNA subunit, TLC1, is predicted to specify the synthesis of the sequence 5′-TGTGTGGGTGTGGTG-3′ if reverse transcription of each repeat is completely processive, yeast telomerase adds imperfect, degenerate repeats with a consensus sequence of 5′-(TG)0-6TGGGTGTG(G)0-1-3′ (Forstemann and Lingner 2001). Thus, telomerase nucleotide addition processivity can be assessed by analyzing the frequency of sequence elements within this consensus. Since almost all telomeric repeats contain the -TGGGTGT- sequence motif, we can analyze reverse transcription of the 5′ portion of the TLC1 template region after the synthesis of this core motif, as well as reverse transcription of the 3′ portion of the template region leading up to the core motif. To study the telomeric sequences, we amplified, cloned, and sequenced telomere VI-R from wild type and the four different rnr1 strains. The distal region of the telomeres has sequences that are divergent because telomerase adds imperfect, degenerate repeats (Forstemann et al. 2000). To ensure that the sequences analyzed were due to telomerase-mediated extension events, only sequences that diverged from bulk telomere sequences were examined.
To assay reverse transcription of the 5′ portion of the TLC1 template region, we analyzed how often the core -TGGGTGT- sequence is followed by a GG dinucleotide, as predicted from the template region of TLC1, to produce either -TGGGTGTGGT- or -TGGG(TG)nTGGGT- repeats (Figure S4A). In a wild-type strain, the core -TGGGTGT- sequence is followed in 52% of all cases by a GG dinucleotide (Figure S4A), similar to what has been previously reported (Forstemann and Lingner 2001). We find that the fraction of repeats that contain the GG dinucleotide in the rnr1-Q288E and rnr1-Q288A mutants is increased to 59% and 61%, respectively, indicating that telomerase processivity for the 5′ region is enhanced in these two mutants (P = 0.018 for rnr1-Q288E and P = 0.003 for rnr1-Q288A, as determined by chi-square tests; Figure S4B). This observation may provide an explanation for the elongated telomeres seen in the rnr1-Q288E and rnr1-Q288A strains. However, no statistically significant difference was observed for the other two rnr1 mutants.
To assess the processivity of the reverse transcription of the 3′ portion of TLC1 template region, we first considered all repeats containing the GG dinucleotide (i.e., -TGGGTGTGGT- repeats). Since the 3′ portion of the template consists of a stretch of CA dinucleotides, multiple alignments are possible for a telomere ending in -TGGT or -TGGTG (Figure 4A). This variable alignment gives rise to the variable number of TG dinucleotides between a GG dinucleotide and the following TGGG motif (Figure 4, A and B). In wild-type cells, we find that there are typically one or two TG dinucleotides between a TGG and the subsequent TGGG motif (Figure 4B), consistent with previous observations (Forstemann and Lingner 2001). In the rnr1-Q288E and the rnr1-Q288A mutants, both of which have increases in the percentage of dGTP and elongated telomeres, there is a shift toward having fewer TG dinucleotides (P = 1 × 10−6 for rnr1-Q288E and P = 4 × 10−10 for rnr1-Q288A, as determined by chi-square tests; Figure 4B). In contrast, the rnr1-R293A and the rnr1-Y285A mutants, which both have decreased percentages of dGTP and shortened telomeres, show an increase in the number TG dinucleotides between a TGG and the following TGGG motif (P = 6 × 10−16 for rnr1-R293A and P = 2 × 10−9 for rnr1-Y285A, as determined by chi-square tests; Figure 4B). If nucleotide addition processivity is low, telomerase would dissociate before reverse transcription proceeds to the next TGGG motif. A cycle of stalling—where telomerase dissociates prematurely, realigns, and reattempts reverse transcription—would increase the number of TG dinucleotides between a TGG and the next TGGG motif. Thus, telomerase in the rnr1-R293A and rnr1-Y285A mutants exhibits reduced nucleotide addition processivity likely due to the low relative levels of dGTP in these mutants.
Figure 4.
Telomerase nucleotide addition processivity is affected by dGTP levels. (A) Schematic illustrating three possible alignments for a telomere ending in -TGGTG with the template region of TLC1. Following reverse transcription and extension of the telomere (with added nucleotides shown in orange), the number of TG dinucleotides between the TGG motif and the following TGGG motif will vary. (B and C) For strains of the indicated genotypes, telomere VI-R was amplified by PCR, cloned, and sequenced. (B) The frequency of having 0, 1, 2, or 3 and higher TG dinucleotides between a TGG and the following TGGG was plotted for each strain. (C) The frequency of having 0, 1, or 2 and higher TG dinucleotides between a TGGG and the following TGGG was plotted for each strain.
Similarly, we can consider all repeats that do not have a GG dinucleotide (i.e., -TGGG(TG)nTGGGT- repeats) and measure the number of TG dinucleotides between a TGGG and the following TGGG. In wild-type cells, one TG dinucleotide usually separates a TGGG and the next TGGG (Figure 4C). Similar to the scenario above, if telomerase nucleotide addition processivity is low, the number of TG dinucleotides between the TGGG motifs would be increased. The rnr1-R293A and rnr1-Y285A mutants, both of which have a reduced percentage of dGTP and shortened telomeres, show a striking increase in the number of TG dinucleotides (P = 1 × 10−5 for rnr1-R293A and P = 2 × 10−23 for rnr1-Y285A, as determined by chi-square tests; Figure 4C), indicating that telomerase processivity is reduced in these strains.
It is formally possible that a change in dGTP levels causes telomerase to preferentially adopt different alignments with the telomere 3′ overhang without altering telomerase processivity. For example, lower dGTP levels may cause telomerase to adopt alignment #3 instead of alignment #1 depicted in Figure 4A, resulting in an increase in TG dinucleotides between TGG and TGGG motifs. However, we do not favor this model because, for both the rnr1-293A and rnr1-Y285A mutants, there is a dramatic increase in the frequency of four or more TG dinucleotides between a TGG motif and the next TGGG motif (Figure S5). Four or more TG dinucleotides can occur only if telomerase dissociates before reverse transcription proceeds to the next TGGG motif because three TG dinucleotides is the maximum possible from a processive telomerase enzyme from the TLC1 sequence (Figure 4A, alignment #3).
It is also possible that these sequence changes are not telomerase-dependent and are instead a consequence of mutations inserted by DNA polymerases. This is not the case as sequencing the rnr1tlc1Δ double mutants described earlier showed that sequence divergence was mostly eliminated in the rnr1tlc1Δ double mutants (Figure S6A) to levels similar to what has been previously observed in telomerase-negative strains (Teixeira et al. 2004; Chang et al. 2011). Furthermore, almost all telomeres analyzed in this study, regardless of the strain, have an identical internal region of 60 bp (Figure S6B), meaning that the sequence changes in the rnr1TLC1 mutants recorded in Figure 4 were confined to the distal end of the telomeres, where telomerase acts. If replication-induced mutations were responsible for the sequence changes in the rnr1TLC1 mutants, these changes would still be observed in the rnr1tlc1Δ mutants, and the changes would also be observed within the internal 60-bp region. Thus, the telomere sequence changes in all four rnr1 mutants are telomerase-dependent.
In this section, we show that all four rnr1 mutants have altered telomerase processivity, even the rnr1-R293A mutant, which shows synergistic effects in the absence of telomerase. We find that telomerase nucleotide addition processivity is influenced by intracellular dGTP levels, with reverse transcription of the 3′ portion of the TLC1 template region being more dramatically affected than the 5′ portion. Altogether, we show that the percentage of dGTP positively correlates both with telomerase processivity and with telomere length.
Characterization of the rnr1Δ deletion
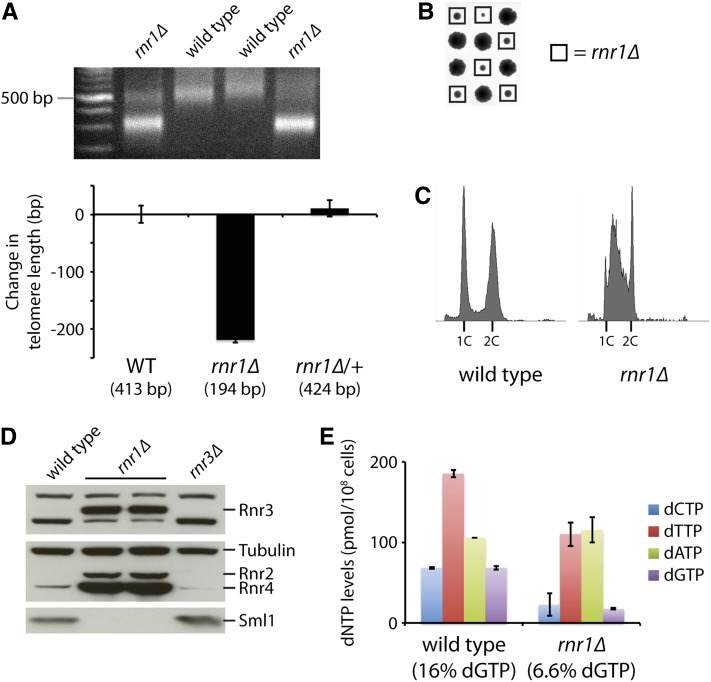
Given our results with the rnr1 point mutants, we decided to examine the effect of an rnr1Δ deletion. Interestingly, while RNR1 is essential in the YNN402 (Elledge and Davis 1990) and W303 backgrounds (I. Sunjevaric and R. Rothstein, unpublished data), an rnr1Δ strain is present in the nonessential gene deletion collection (Giaever et al. 2002), which is in the BY4741 strain background, a derivative of the S288C background. This rnr1Δ strain has been reported to have really short telomeres, >200 bp shorter than those in wild-type strains (Gatbonton et al. 2006), but considering the discrepancy in the reported viability of rnr1Δ mutants, we decided to validate the reported phenotypes ourselves. We first confirmed that the rnr1Δ mutant from the deletion collection has really short telomeres (data not shown). In this strain, the RNR1 gene has been replaced with the kanMX4 cassette, which provides resistance to the drug geneticin (also known as G418). We backcrossed the rnr1Δ strain twice to a BY4741 wild-type strain and found that the short telomere phenotype always cosegregated with resistance to G418 and slow growth (Figure 5, A and B). We then verified the location of the kanMX4 cassette by PCR amplification of the junctions between the cassette and locations upstream and downstream of RNR1 (data not shown).
Figure 5.
rnr1Δ mutants have shortened telomeres due to reduced dGTP levels. (A) An rnr1Δ mutant from the yeast gene deletion collection was backcrossed to a wild-type strain (BY4741) twice. The resulting wild-type and rnr1Δ progeny strains, along with a heterozygous rnr1Δ/RNR1 diploid, were assayed for telomere length by Y′ telomere PCR after being passaged for at least 100 generations (a representative gel is shown). The change in telomere length, compared to wild-type telomere length, was quantified and plotted. Mean ± SE is plotted for at least four independent isolates (two for the rnr1Δ/RNR1 diploid). (B) Tetrad analysis reveals that an rnr1Δ strain exhibits slow growth. Each column of four colonies is a single tetrad derived from the sporulation of an rnr1Δ/RNR1 diploid followed by the separation of the four haploid spores by micromanipulation. (C) Flow cytometry histograms for the indicated strains derived from B. (D) Wild-type, rnr1Δ, and rnr3Δ strains were assayed for Rnr3, Rnr2, Rnr4, and Sml1 protein levels by protein blot analysis. Tubulin levels were also assayed as a loading control. (E) dNTP pools in the wild-type and rnr1Δ strains were measured. Data are represented as mean ± SE. dGTP levels, as a percentage of total dNTPs, are indicated for each strain.
Flow cytometric analysis revealed defects in cell-cycle progression in the rnr1Δ mutant, with many cells delayed in S phase (Figure 5C). We also find that there is upregulation of Rnr2, Rnr4, and most significantly, Rnr3 (Figure 5D). Rnr3 is a minor isoform of the large subunit of RNR that is expressed only following DNA damage or replication blocks in response to an increased need for dNTPs (Elledge and Davis 1990). Furthermore, Sml1 is degraded in rnr1Δ strains (Figure 5D), which normally occurs during S phase or in response to DNA damage (Zhao et al. 2001). We suspect that the changes in RNR and Sml1 levels are likely responsible for keeping the cells viable. However, we are still uncertain why a deletion of RNR1 is viable in the BY4741 background, but lethal in the YNN402 and W303 backgrounds. Interestingly, we find that the difference in viability is due to one genetic locus, which is still unknown (see File S1, Supporting information regarding the viability of rnr1Δ).
We next examined the levels of the four dNTPs in the rnr1Δ mutant. While the levels of dATP appear similar between wild-type cells and rnr1Δ mutants, the levels of the other three dNTPs are substantially reduced (Figure 5E). In particular, the percentage of dGTP is reduced from 16% in wild-type cells to 6.6% in the rnr1Δ strain (Figure 5E). This reduction in dGTP, combined with the overall reduction in dNTPs, likely contributes significantly to the drastic shortening of telomeres seen in rnr1Δ mutants (Figure 5A).
Human telomerase activity is dramatically affected by changes in dGTP concentration
Since our results suggest that telomerase and telomere length are extremely sensitive to dGTP levels, we decided to test whether our findings in yeast are evolutionarily conserved. Previous in vitro studies of ciliate (Hammond and Cech 1997, 1998; Hardy et al. 2001) and mammalian (Morin 1989; Maine et al. 1999) telomerase suggested that telomerase activity is stimulated by dGTP. However, these studies did not address the effect of separately changing the concentration of all four dNTPs. In light of our yeast in vivo observations, we decided to measure human telomerase activity, using the Telospot assay (Cristofari and Lingner 2006), while systematically titrating all four dNTPs. In this assay, both the telomerase RNA subunit, hTR, and the protein catalytic subunit, hTERT, are strongly overexpressed after transient transfection, yielding a situation referred to as “super-telomerase.” Crude super-telomerase extract is incubated with a telomeric (TTAGGG)3 primer with varying concentrations of dNTPs. A small fraction of the reaction is directly spotted onto a nylon membrane, which is then probed with a randomly radiolabeled telomeric probe. Since mammalian and S. cerevisiae dNTP pools are similar in concentration and in terms of the ratio of the four dNTPs (Traut 1994; Sabouri et al. 2008; Nick McElhinny et al. 2010), we used the yeast dNTP concentrations determined from the wild-type strain used in Figure 3A as the starting point for our assays (Figure S7A). We maintained three of the dNTPs at these “physiological concentrations” while varying the fourth. Titration of dCTP, dATP, or dTTP does not dramatically affect telomerase activity (Figure S7, A and B). However, consistent with previous results (Maine et al. 1999), alteration of dGTP had a striking effect on telomerase activity (Figure S7, A and B). Human telomerase activity is markedly reduced when dGTP concentrations are lowered, while activity is greatly increased even with modest increases in dGTP concentration.
An increase in telomerase activity in the Telospot assay is likely due to processive addition of telomere repeats to the primers to yield long extended products, but it could also result from many primers extended only a short distance. To differentiate between these two scenarios, we repeated the reactions where dGTP concentration was varied using a 5′-biotinylated (TTAGGG)3 primer and resolved the products on a polyacrylamide gel after purification with streptavidin-coated beads. The DNA was transferred onto a nylon membrane and probed as in a standard Telospot assay (Figure S7C). Consistent with previous observations (Maine et al. 1999), processive telomerase activity, as measured by the increase in size of the fragments, positively correlates with dGTP concentration. These results indicate that the specific effect of dGTP levels on telomerase activity is an evolutionarily conserved feature.
Discussion
In this study, we examined the effect of changing intracellular dNTP levels, including unbalancing the four dNTPs, on telomere length homeostasis in S. cerevisiae. When the ratio of the four dNTPs is maintained, we find that telomere length decreases modestly when total dNTP levels are reduced, while there is no significant change in telomere length upon increasing total dNTP levels. However, we demonstrate that altering the ratio of the dNTPs has a much more pronounced effect on telomere length homeostasis. Specifically, we show that both telomerase nucleotide addition processivity and telomere length positively correlate with dGTP levels in vivo.
Reduction of dNTPs leads to shorter telomeres but increase of dNTPs does not significantly affect telomere length
Although previous studies hinted at a connection between dNTP pools and telomere length homeostasis, our work is the first to document their precise relationship. We find that changing the total levels of dNTPs, without altering the ratio of the dNTPs, leads to a modest change in telomere length. Deletion of DUN1 leads to a twofold reduction in dNTPs and an ∼50-bp reduction in telomere length (Figure 1A and Figure S1). However, a sml1Δ crt1Δ double mutant has a fourfold increase in dNTP levels but an increase in telomere length of <20 bp (Figure 1C and Figure S2A). Thus, an excess of dNTPs results in a rather minimal increase in telomere length. We were unable to probe the telomere length effects of a reduction of dNTP levels greater than twofold without altering the ratio of the four dNTPs because a mutant with such low levels has not been reported. Presumably, such a mutant would be inviable due to insufficient levels of dNTPs required for DNA synthesis.
Mec1 mediates telomere length homeostasis by regulating dNTP levels
Previous work has shown that mec1-21 mutants have shortened telomeres that can be rescued by deletion of SML1 (Ritchie et al. 1999). Given that mec1-21, dun1Δ, and dun1Δ mec1-21 mutants all have similar dNTP levels (Fasullo et al. 2010), we asked whether the shortened telomeres in the mec1-21 mutant are due to reduced activation of Dun1. By epistasis analysis, we show this to be the case, with a dun1Δ mec1-21 double mutant having the same telomere length as each of the single mutants (Figure 2). However, Mec1 has functions at the telomere that are Dun1-independent. While mec1-21 tel1Δ double mutants senesce, similar to a telomerase-negative strain (Ritchie et al. 1999), dun1Δ tel1Δ mutants fail to senesce, despite repeated subculturings (data not shown). Thus, while Mec1 affects telomere length homeostasis through Dun1-mediated regulation of dNTP pools, Mec1 also has Dun1-independent functions at the telomere.
Intracellular dGTP levels affect telomere length homeostasis by altering telomerase nucleotide addition processivity
The most remarkable finding from our work is the strong dependence of telomere length homeostasis and telomerase activity on the levels of dGTP in the cell. Specifically, we find that the length of yeast telomeres and nucleotide addition processivity of telomerase positively correlate with intracellular dGTP levels (Figures 3 and 4).
Consistent with our observations, yeast telomerase mutants that alter nucleotide addition processivity, as measured in vitro, positively correlate with the in vivo length of the telomeres (Peng et al. 2001), and this processivity is enhanced by increasing dGTP concentrations (Bosoy and Lue 2004). However, these in vitro telomerase assays examined only the processivity of nucleotide addition using the 5′ portion of the TLC1 template region, whereas we were able to examine in vivo both the 3′ and 5′ portions and have found that reverse transcription of the 3′ portion is more dramatically affected by changes in dGTP levels.
Telomerase enzymes can also be characterized by their ability to add multiple repeats before dissociating (i.e., their repeat addition processivity). Yeast telomerase is generally nonprocessive at adding repeats, both in vitro (Cohn and Blackburn 1995) and in vivo (Chang et al. 2007). However, yeast telomerase has the ability to processively elongate critically short telomeres in vivo (Chang et al. 2007), and limited repeat addition processivity can be observed in vitro by increasing dGTP concentration (Bosoy and Lue 2004).
In agreement with these findings in budding yeast, previous studies have also observed dGTP-dependent stimulation of human telomerase activity in vitro (Morin 1989; Maine et al. 1999), and by careful titration of all four dNTPs, we show that this effect is specific to dGTP (Figure S7C). Similar in vitro studies of endogenous Euplotes telomerase and recombinant Tetrahymena telomerase have also revealed that it is the binding of dGTP to telomerase that stimulates its repeat addition processivity (Hammond and Cech 1997, 1998; Hardy et al. 2001). Interestingly, while we find that the percentage of total dNTPs that is dGTP is important in vivo in yeast (Figure 4), it is the absolute concentration of dGTP, independent of the levels of the other three dNTPs, which is important for in vitro human telomerase activity (Figure S7). It is noteworthy that the biochemical characteristics of yeast, ciliate, and human telomerases are quite different. For example, they show differences in processivity and associate with different complements of proteins, and their RNA templates differ vastly (Mason et al. 2011). Thus, it is quite remarkable that the importance of dGTP levels on telomerase activity is highly conserved, even if the precise mechanism may differ in different species.
Our results indicate that intracellular dGTP levels are rate-limiting for both yeast and human telomerase activity. dNTP levels are most likely optimized for the DNA synthesis machinery. Low levels of dNTPs cause DNA replication fork stalling (Desany et al. 1998) while high levels of dNTPs result in an increase in mutation rate (Reichard 1988; Chabes et al. 2003). It has also been shown that even mild dNTP pool imbalances are mutagenic (Kumar et al. 2010). However, our work suggests that telomere length homeostasis may also impose selective pressure on optimal intracellular dGTP levels. Indeed, of the four dNTPs, dGTP levels are kept the lowest in both yeast (Chabes et al. 2003) and mammalian cells (Traut 1994), perhaps reflecting an evolutionarily conserved mechanism to regulate telomerase activity and telomere length homeostasis.
Finally, our findings may provide new strategies to regulate telomerase activity therapeutically. For example, telomerase is repressed in most human somatic cells but is expressed in ∼85% of cancers (Shay and Bacchetti 1997), and telomerase has been viewed as an ideal target to inhibit the growth of a wide range of tumors. Identifying agents that reduce intracellular dGTP levels may be effective in limiting the proliferation of cancer cells. Furthermore, several human diseases are associated with shortened telomeres. Individuals born with reduced telomerase activity have short telomeres, leading to telomere dysfunction in highly proliferative cells (Armanios 2009). Indeed, many of these individuals are haploinsufficient for telomerase and have shortened life spans, suggesting that full telomerase activity is important in preventing these diseases. Perhaps elevating the levels of dGTP to increase telomerase activity will be effective in treating these individuals. Thus, it will be of significant interest to find ways to modulate intracellular dGTP levels as a mechanism to regulate telomerase activity.
Acknowledgments
We thank Peter Thorpe for constructive comments on the manuscript; Stephen Elledge for providing strains; Dinesh Kumar for help in constructing yeast strains; Ivana Sunjevaric for allowing us to cite her unpublished data; and Gaël Cristofari for advice on the Telospot assays. S.S. was supported by a stipend from the Wenner-Gren Foundations. M.C. was supported by a Terry Fox Foundation Fellowship Award. This work was supported by funds from the Swiss National Science Foundation and a European Research Council advanced investigator grant (to J.L.); the Swedish Foundation for Strategic Research, the Swedish Research Council, and the Swedish Cancer Society (A.C.); and National Institutes of Health grants GM50237 and GM67055 (to R.R.).
Footnotes
Communicating editor: N. M. Hollingsworth
Literature Cited
- Adams A. K., Holm C., 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 4614–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M., 2009. Syndromes of telomere shortening. Annu. Rev. Genomics Hum. Genet. 10: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnerić M., Lingner J., 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 8: 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosoy D., Lue N. F., 2004. Yeast telomerase is capable of limited repeat addition processivity. Nucleic Acids Res. 32: 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A., Domkin V., Thelander L., 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274: 36679–36683. [DOI] [PubMed] [Google Scholar]
- Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., et al. , 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401. [DOI] [PubMed] [Google Scholar]
- Chang M., Arneric M., Lingner J., 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21: 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Dittmar J. C., Rothstein R., 2011. Long telomeres are preferentially extended during recombination-mediated telomere maintenance. Nat. Struct. Mol. Biol. 18: 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M., Blackburn E. H., 1995. Telomerase in yeast. Science 269: 396–400. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Lingner J., 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin R. A., Kalashnikova T. I., Chahwan C., McDonald W. H., Wohlschlegel J., et al. , 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23: 483–496. [DOI] [PubMed] [Google Scholar]
- Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J., 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12: 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W., 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4: 740–751. [DOI] [PubMed] [Google Scholar]
- Fasullo M., Tsaponina O., Sun M., Chabes A., 2010. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 38: 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Funk W. D., Wang S. S., Weinrich S. L., Avilion A. A., et al. , 1995. The RNA component of human telomerase. Science 269: 1236–1241. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Lingner J., 2001. Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol. 21: 7277–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K., Hoss M., Lingner J., 2000. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 28: 2690–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., et al. , 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., et al. , 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337. [DOI] [PubMed] [Google Scholar]
- Hammond P. W., Cech T. R., 1997. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 25: 3698–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P. W., Cech T. R., 1998. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry 37: 5162–5172. [DOI] [PubMed] [Google Scholar]
- Hardy C. D., Schultz C. S., Collins K., 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276: 4863–4871. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Futcher A. B., Greider C. W., 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J., 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Jain D., Cooper J. P., 2010. Telomeric strategies: means to an end. Annu. Rev. Genet. 44: 243–269. [DOI] [PubMed] [Google Scholar]
- Kumar D., Viberg J., Nilsson A. K., Chabes A., 2010. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 38: 3975–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. D., Elledge S. J., 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 20: 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. D., Wang J., Stubbe J., Elledge S. J., 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell 32: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., et al. , 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567. [DOI] [PubMed] [Google Scholar]
- Lundblad V., Szostak J. W., 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Maine I. P., Chen S. F., Windle B., 1999. Effect of dGTP concentration on human and CHO telomerase. Biochemistry 38: 15325–15332. [DOI] [PubMed] [Google Scholar]
- Mason M., Schuller A., Skordalakes E., 2011. Telomerase structure function. Curr. Opin. Struct. Biol. 21: 92–100. [DOI] [PubMed] [Google Scholar]
- Meyne J., Ratliff R. L., Moyzis R. K., 1989. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 86: 7049–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin G. B., 1989. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59: 521–529. [DOI] [PubMed] [Google Scholar]
- Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., et al. , 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundstrom E. B., et al. , 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA 107: 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Ma E., Marcand S., 2006. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae. Genetics 172: 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mian I. S., Lue N. F., 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Peter M., Gartner A., Horecka J., Ammerer G., Herskowitz I., 1993. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73: 747–760. [DOI] [PubMed] [Google Scholar]
- Reichard P., 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57: 349–374. [DOI] [PubMed] [Google Scholar]
- Ritchie K. B., Mallory J. C., Petes T. D., 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N., Viberg J., Goyal D. K., Johansson E., Chabes A., 2008. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 36: 5660–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W., Bacchetti S., 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33: 787–791. [DOI] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E., 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409. [DOI] [PubMed] [Google Scholar]
- Tang H. M., Siu K. L., Wong C. M., Jin D. Y., 2009. Loss of yeast peroxiredoxin Tsa1p induces genome instability through activation of the DNA damage checkpoint and elevation of dNTP levels. PLoS Genet. 5: e1000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. T., Arneric M., Sperisen P., Lingner J., 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335. [DOI] [PubMed] [Google Scholar]
- Traut T. W., 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140: 1–22. [DOI] [PubMed] [Google Scholar]
- Tsaponina O., Barsoum E., Astrom S. U., Chabes A., 2011. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 7: e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Huang M., 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol. Cell. Biol. 28: 7156–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Bradley J. D., Attardi L. D., Blackburn E. H., 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller E. G., Rothstein R., 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340. [DOI] [PubMed] [Google Scholar]
- Zhao X., Chabes A., Domkin V., Thelander L., Rothstein R., 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]