Figure 5.
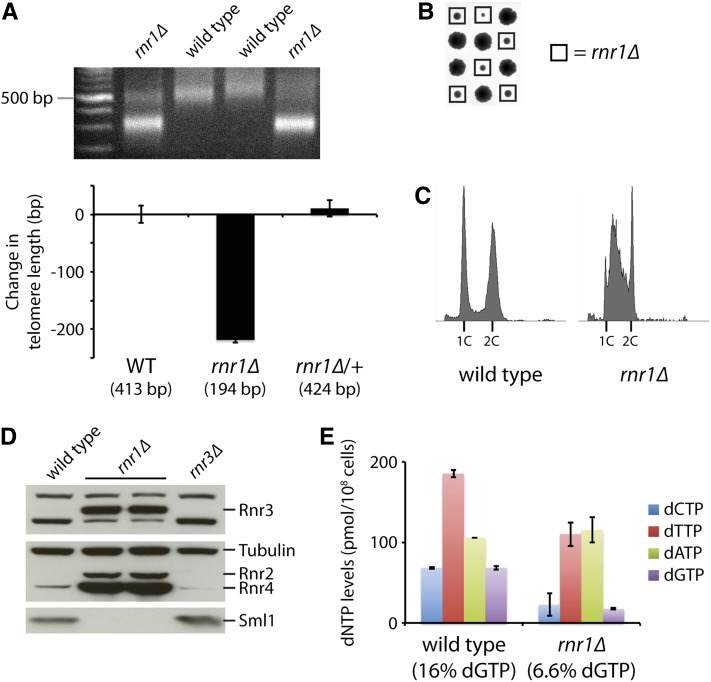
rnr1Δ mutants have shortened telomeres due to reduced dGTP levels. (A) An rnr1Δ mutant from the yeast gene deletion collection was backcrossed to a wild-type strain (BY4741) twice. The resulting wild-type and rnr1Δ progeny strains, along with a heterozygous rnr1Δ/RNR1 diploid, were assayed for telomere length by Y′ telomere PCR after being passaged for at least 100 generations (a representative gel is shown). The change in telomere length, compared to wild-type telomere length, was quantified and plotted. Mean ± SE is plotted for at least four independent isolates (two for the rnr1Δ/RNR1 diploid). (B) Tetrad analysis reveals that an rnr1Δ strain exhibits slow growth. Each column of four colonies is a single tetrad derived from the sporulation of an rnr1Δ/RNR1 diploid followed by the separation of the four haploid spores by micromanipulation. (C) Flow cytometry histograms for the indicated strains derived from B. (D) Wild-type, rnr1Δ, and rnr3Δ strains were assayed for Rnr3, Rnr2, Rnr4, and Sml1 protein levels by protein blot analysis. Tubulin levels were also assayed as a loading control. (E) dNTP pools in the wild-type and rnr1Δ strains were measured. Data are represented as mean ± SE. dGTP levels, as a percentage of total dNTPs, are indicated for each strain.