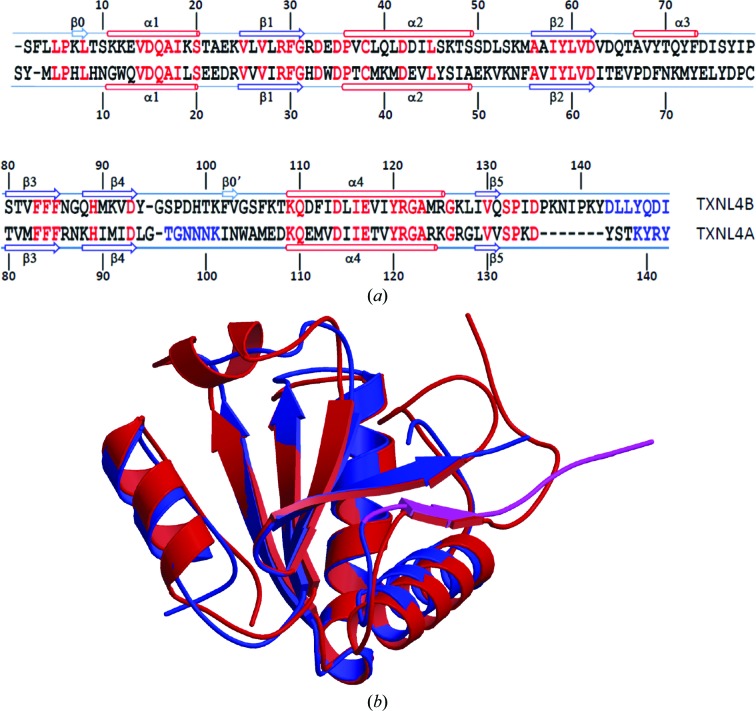
Figure 4.
Comparison between TXNL4A and TXNL4B. (a) Sequence alignment based on structure superposition by MUSTANG. Single-letter amino-acid codes are used. Red indicates identical residues in the two proteins. Rather than structural alignment, blue indicates amino acids that were not located in the electron-density maps. Secondary-structure elements as assigned by STRIDE (Heinig & Frishman, 2004 ▶) are shown above the sequence for TXNL4B and below the sequence for TXNL4A. β-Strands are shown as arrows and α-helices are shown as cylinders. They are numbered from the N-terminus of the proteins, except for two strands in TXNL4B that consist of only two residues, which are numbered separately. These two short strands are not shown as strands in (b). (b) Structure comparison between TXNL4A and TXNL4B. Each structure is represented as a ribbon diagram, with monomer A of TXNL4B shown in red and TXNL4A shown in blue, except for residues Leu129–Thr138 (shown in magenta) which are within the functionally important 14-residue tail of TXNL4A, the removal of which is known to result in a dominant negative form of the protein.