Table 1. Crystallographic statistics for PaIMPDH.
Values in parentheses are for the highest resolution shell.
| Space group | I4 |
| Unit-cell parameters (Å) | a = b = 115.5, c = 56.4 |
| Resolution (Å) | 19.1–2.25 (2.37–2.25) |
| No. of reflections recorded | 86989 (12454) |
| Unique reflections | 16612 (2476) |
| Completeness (%) | 94.6 (97.0) |
| Multiplicity | 5.2 (5.0) |
| 〈I/σ(I)〉 | 16.3 (4.3) |
| Wilson B (Å2) | 29.3 |
| No. of residues | 293 |
| No. of waters | 117 |
| R merge † (%) | 9.1 (48.9) |
| R work ‡ (%) | 14.8 |
| R free § (%) | 19.0 |
| Average B factor (Å2) | |
| Protein | 33.7 |
| Waters | 38.5 |
| Chloride | 72.1 |
| Cruickshank DPI¶ (Å) | 0.2 |
| Ramachandran plot | |
| Most favoured (%) | 97.3 |
| Additional allowed (%) | 2.0 |
| Outliers (%) | 0.7 |
| R.m.s.d. from ideal values†† | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.5 |
R
merge = 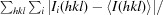
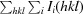 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
R
work = 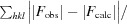
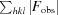 , where F
obs is the observed structure factor and F
calc is the calculated structure factor.
, where F
obs is the observed structure factor and F
calc is the calculated structure factor.
R free is the same as R cryst except that it was calculated with a subset (5%) of data that were excluded from the refinement calculations.
Diffraction precision index (Cruickshank, 1999 ▶).
Engh & Huber (1991 ▶).
