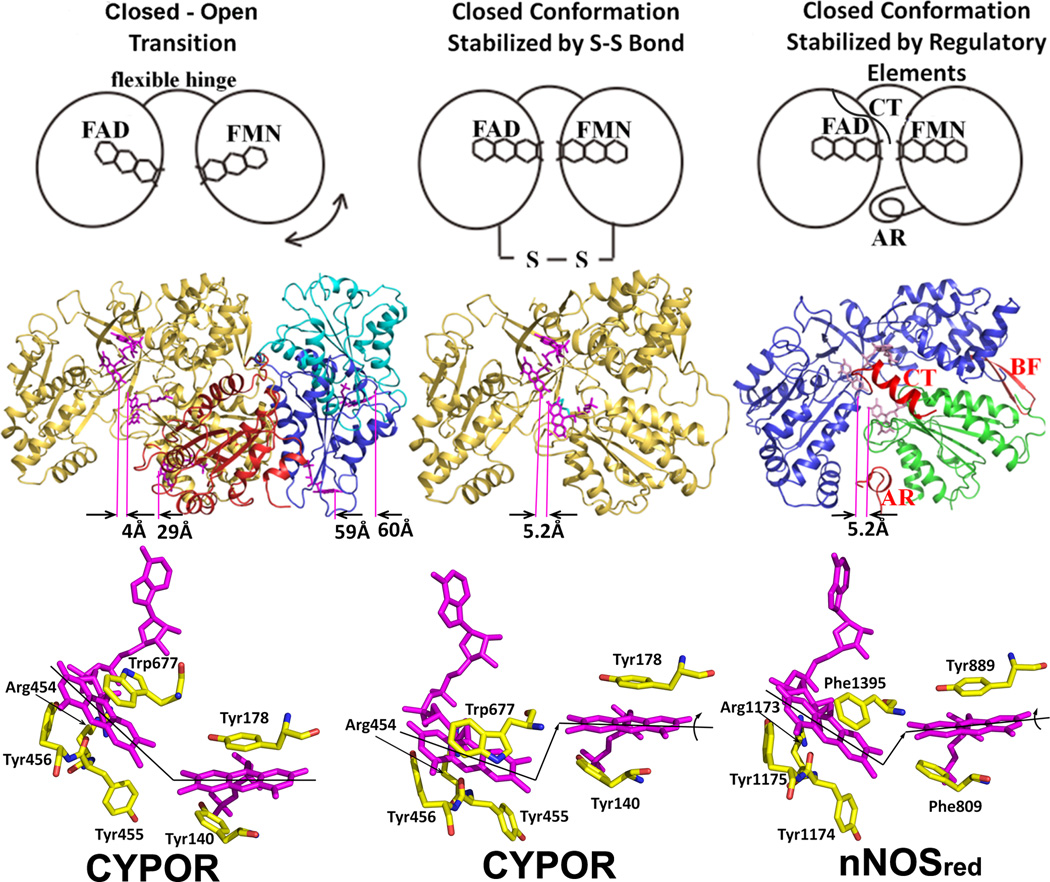
Figure 5.
The closed and open conformations of CYPOR (left and middle panels) and nNOSred (right panel). FAD and FMN are shown with stick models in magenta. In the middle row, ribbon diagrams for CYPOR and nNOS red structures are shown. Distances between FAD and FMN (between arrows indicating C8-C8 carbon atoms pointed by purple lines) are shown in Å in the middle row. The left panel shows relative orientations of FMN domain with respect to the FAD domain observed in WT (gold, PDB code: 1AMO) and three open conformations of the linker-deleted mutant (PDB code: 3FJO), Mol A (red), Mol B (blue) and Mol C (cyan) are shown (see ref [54] for details). In the middle panel, the engineered S-S bond (green) is shown in the closed-form mutant of CYPOR (PDB code: 3OJW, middle panel, see ref. [56] for details); and nNOSred containing the regulatory elements, AR, CT and BF of NOSred (PDB code: 1TLL, see ref. [42] for details) is shown in the right panel. In the bottom row, relative orientations of FMN and FAD are schematically shown by lines representing the isoalloxazine planes. In the wild type structure (left panel), the two planes make ~135 and form a continuous ribbon and aligned, while in the disulfide-engineered mutant the two planes are not aligned (middle panel, bottom). It is not clear whether the difference in the FMN-FAD distances observed in the closed form of wild type CYPOR (4.0 Å) and nNOSred (5.2 Å) and the slight shift and rotation of the two flavin planes observed in the NOSred structure (right panel, bottom). However, it is possible that, in NOS, CaM may unlock the closed conformation to make the distance and orientation between FAD and FMN more favorable for efficient ET between them.