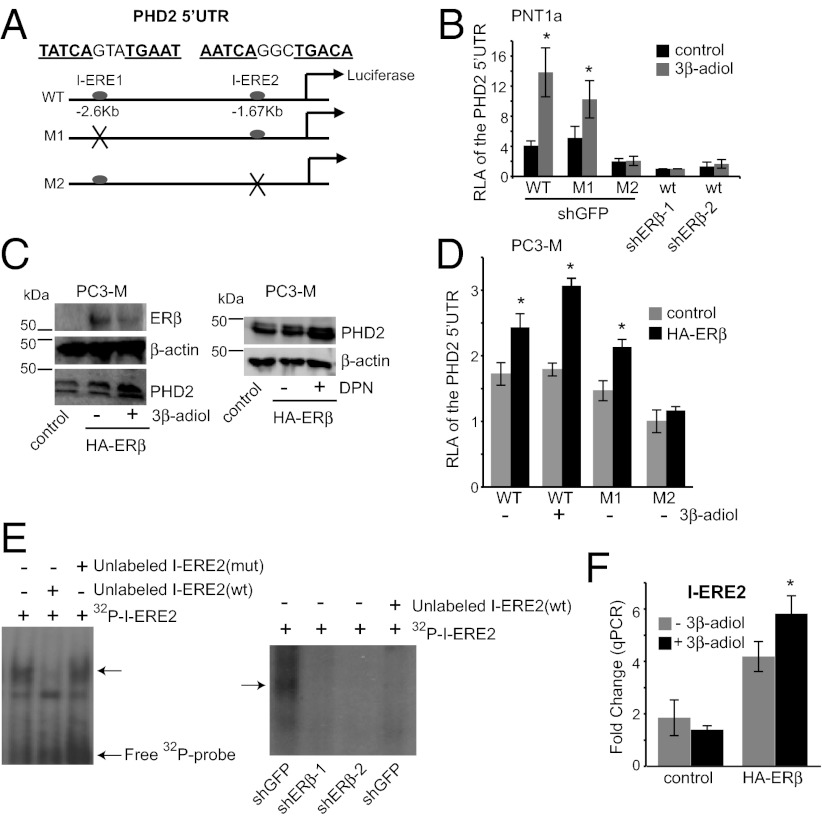
Fig. 2.
ERβ regulates PHD2 transcription via a unique ERE, located in the 5′ UTR of the PHD2 gene. (A) Schematic of the wild-type and mutant luciferase reporter constructs used. A 927-bp fragment (−2597 bp to −1670 bp), containing two variant EREs (I-ERE1: TATCAgtaTGAAT and I-ERE2: AATCAggcTGACA), was cloned into a pGL3-promoter construct (WT). M1 represents the mutated version of I-ERE1 (TAAAAGTAAAAAT), whereas M2 represents the mutated version of I-ERE2 (AAAAAGGCAAACA). These constructs were used for subsequent experiments. (B) PNT1a cells were transfected with WT, M1, or M2 luciferase constructs. ERβ-ablated cells (shERβ-1 and shERβ-2) were transfected with the WT construct together with Renilla for normalization. Cells were subsequently incubated for 20 h in the absence or presence of 3β-adiol. Note that the relative luciferase activity (RLA) of the M2 construct is significantly less than that of the WT and M1 constructs and comparable to that of the wild-type construct in ERβ-ablated cells in the absence or presence of 3β-adiol (*P < 0.05). However, only the wild-type and M1 constructs exhibit ligand responsiveness (two- to threefold increase in luciferase activity). (C) Ligand-activated ERβ promotes PHD2 expression in PC3-M cells. PC3-M cells, which have an undetectable level of ERβ, were infected with an empty vector or an HA-ERβ expression vector. ERβ expressing cells (lanes 2 and 3) were incubated with DMSO (control), 3β-adiol (5 µM), or DPN (30 nM) for 24 h and harvested for immunoblotting. ERβ expression alone increases PHD2 expression and this effect is enhanced by either 3β-adiol or DPN. (D) Analyses of ERE-luciferase activity in PC3-M cells. PC3-M cells that express an HA-ERβ or an empty vector (control) were transfected with either WT (±3β-adiol), M1, or M2 constructs together with Renilla. Expression of HA-ERβ increases luciferase activity, which is enhanced by 3β-adiol (*P < 0.05). This enhancement is not apparent with M2. (E) EMSA assays to detect ERβ–DNA complex formation in vitro. Nuclear extracts containing ERβ from 3β-adiol–treated PNT1a cells or ERβ-ablated cells were incubated with 32P-labeled I-ERE2 (WT) in the absence or presence of unlabeled competitors. A distinct protein–DNA complex was detected (arrow) in PNT1a cells but not in ERβ-ablated cells (shERβ-1 and shERβ-2) and this complex was displaced by unlabeled I-ERE2 (WT) but not I-ERE2 (mut) confirming its specificity. (F) ChIP analysis of HA-ERβ–expressing PC3-M cells. PC3-M (+HA-ERβ) or PC3-M cells (empty vector) were incubated in the absence or presence of 3β-adiol for 48 h. Either an HA antibody or IgG (control) were used to precipitate the chromatin. Enrichment on the ERE2 locus by HA was normalized to IgG by qPCR. I-ERE2 mRNA levels exhibit a fourfold induction in HA-ERβ–expressing cells and a sixfold induction upon treatment with 3β-adiol. In contrast, no enrichment in I-ERE2 mRNA is apparent in control PC3-M cells (*P < 0.05) under the same conditions.