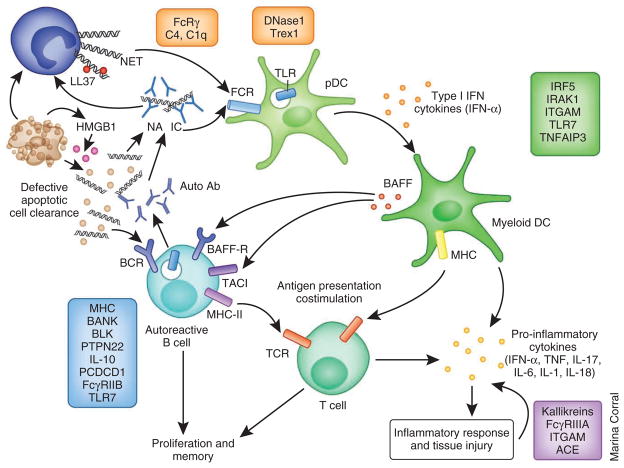
Figure 2.
The role of SLE risk alleles in the pathogenesis of SLE. A panoply of genetic variations has been linked to SLE susceptibility. Polymorphisms in genes involved in the immune clearance of apoptotic particles and nucleic-acid–containing immune complexes (clearance functions, with examples shown in orange) may induce the enhanced activation of pDCs and autoreactive B cells, leading to the production of type I IFN and the expansion of autoreactive effector cells, respectively. Polymorphisms in genes involved in innate immunity (with examples shown in green) regulate the induction of, as well as the response to, type I IFN. Abnormal function of innate immune cells in turn activates the adaptive immune system. Both the innate and adaptive immune systems contribute to the inflammatory response and tissue damage. A third major group of polymorphic genes is involved in ligand recognition, receptor signaling and other immunological functions of adaptive immune cells (with examples shown in blue). Dysregulation of the adaptive immune system results in loss of tolerance and the production of autoantibodies, which in turn bind to nuclear antigens and activate innate immune cells, completing a positive feedback loop that amplifies the pathogenic processes in SLE. Polymorphic alleles may also influence the severity of organ damage (with examples shown in purple). Ab, antibody; IRAK1, interleukin-1 receptor-associated kinase 1; ITGAM, integrin α M; TNFAIP3, tumor necrosis factor, α-induced protein 3; BANK, B cell scaffold protein with ankyrin repeats; BLK, B lymphoid tyrosine kinase; PCDCD1, programmed cell death 1; NA, nucleic acid; IC, immune complex; ACE, angiotensin I converting enzyme (peptidyl-dipeptidase A) 1.