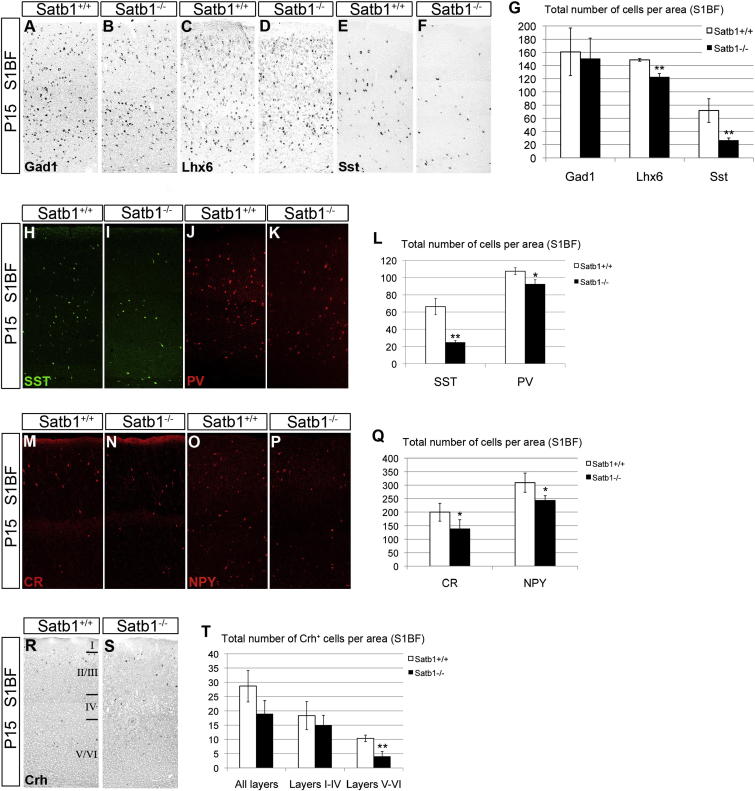
Figure 5.
Satb1 Deletion Results in Impaired Differentiation of SST+ Cortical Interneurons
(A–F) In situ hybridization histochemistry of P15 cortical sections from wild-type (A, C, and E) and Satb1-deficient (B, D, and F) mice with riboprobes specific for Gad1 (A and B), Lhx6 (C and D), and Sst (E and F).
(G) Quantification of Gad1-, Lhx6-, and Sst-expressing cells in the somatosensory cortex (S1BF, bregma 0.38 mm) of wild-type (white bars) and Satb1-deficient mice (black bars).
(H–K) Immunofluorescence of P15 cortical sections from wild-type (H and J) and Satb1-deficient (I and K) mice with antibodies specific for SST (H and I) and PV (J and K).
(L) Quantification of SST+ and PV+ cells in the somatosensory cortex (S1BF, bregma 0.38 mm) of wild-type (white bars) and Satb1-deficient mice (black bars).
(M–P) immunofluorescence of P15 cortical sections from wild-type (M and O) and Satb1-deficient (N and P) mice with antibodies specific for CR (M and N) or NPY (O and P).
(Q) Percentages of CR+ and NPY+ cells in wild-type (white bars) and Satb1-deficient cortex (black bars).
(R and S) In situ hybridization histochemistry of P15 cortical sections from wild-type (R) and Satb1 mutant (S) mice with a Crh-specific riboprobe.
(T) Quantification of CRH+ neurons in the layers of the somatosensory cortex (S1BF, bregma 0.38 mm) of wild-type (white bars) and Satb1-deficient mice (black bars).
n = 3 animals per each interneuron marker analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 for each test (Student’s homoscedastic t test with two-tailed distibution). See also Figures S4, S5, and S7.