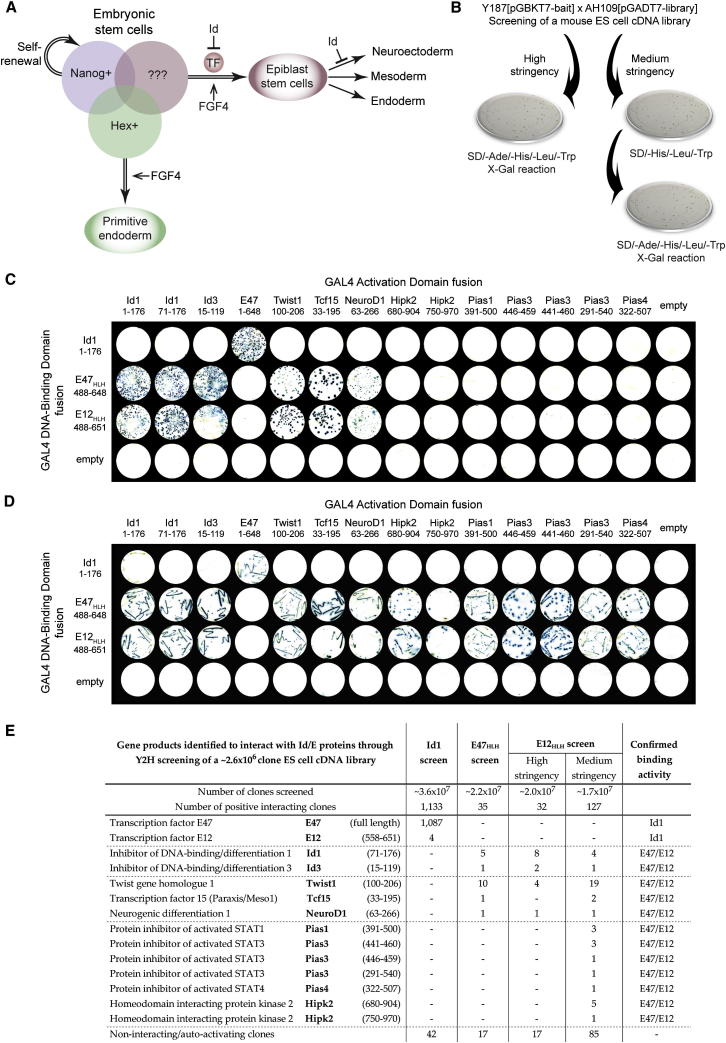
Figure 1.
A Y2H Screen to Identify Transcription Factors Regulated by Id1 in ES Cells
(A) FGF signaling primes pluripotent cells for differentiation. Id1 inhibits subsequent transitions: first to epiblast and then to neuroepithelium. We hypothesize that unknown Id-regulated transcription factors are expressed in epiblast-primed cells.
(B) A Y2H library screen was performed by mating the Y187 yeast strain carrying the pGBKT7-bait vector (Id1, E47HLH, or E12HLH) with the AH109 yeast strain carrying the pGADT7-library vector (mouse ESC cDNA library). For high-stringency selection, positive interactors were determined by growth of mated yeast on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction (indicating activation of ADE2, HIS1 and LacZ reporter genes). For medium-stringency selection, positive interactions were determined by initial growth of mated yeast on SD/-His/-Leu/-Trp media (indicating activation of HIS1 reporter gene), with subsequent growth on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction (indicating activation of ADE2, HIS1 and LacZ reporter genes).
(C) High-stringency Y2H analysis of interactions among Id1, E47HLH, and E12HLH, and genes identified through library screens; positive interactors are determined by growth on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction.
(D) Medium-stringency Y2H analysis of interactions among Id1, E47HLH, and E12HLH, and genes identified through library screens; positive interactions are determined by initial growth on SD/-His/-Leu/-Trp media with subsequent growth on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction.
(E) The genes identified as pGADT7 vector inserts that interact with Id1, E47HLH, or E12HLH at the protein level within yeast are shown along with the number of times clones were identified in each screen, and whether each gene was confirmed as a true binding partner of Id1 or E47/E12 by subsequent analysis. The total numbers of noninteracting and autoactivating inserts identified are also shown.