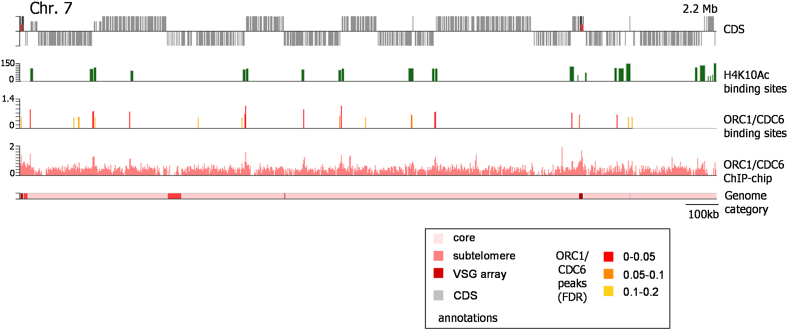
Figure 1.
Mapping ORC1/CDC6 Binding in the T. brucei Nuclear Genome
To map T. brucei ORC1/CDC6 localization sites, ChIP was performed and the recovered DNA cohybridized with input DNA to a microarray; data are shown for chromosome (Chr.) 7. The bottom panel delineates the chromosome into “core,” “subtelomere-proximal,” and “VSG array” sequences (see text for details, and inset for coloring). The location of CDSs (gray boxes) along the chromosome is shown in the top panel; genes above the line are transcribed toward the right, and those below are transcribed toward the left. Enrichment of ORC1/CDC6-bound DNA relative to the input is shown in the second bottom panel; values are plotted as the log2 ratio (y axis) of sample/input and were calculated over a 500 bp sliding window. In the panel above, predicted ORC1/CDC6 binding sites are identified as “peaks,” which are shown by vertical lines colored to indicate the likelihood of being an ORC1/CDC6 binding site based on three categories of FDR: red indicates the highest confidence (FDR ≤0.05), and orange and yellow decreasing confidence (FDRs of 0.05–0.1 and 0.1–0.2, respectively). In the second top panel, sites of H4K10Ac localization are indicated as green vertical lines; these data are derived from ChIP-seq data of Siegel et al. (2009), identifying positions of likely transcription start sites, and are represented as log2 values (y axis) in 250 bp windows, with the width of the lines indicating the areas of the chromosome covered by the modified histone. See also Figure S1.