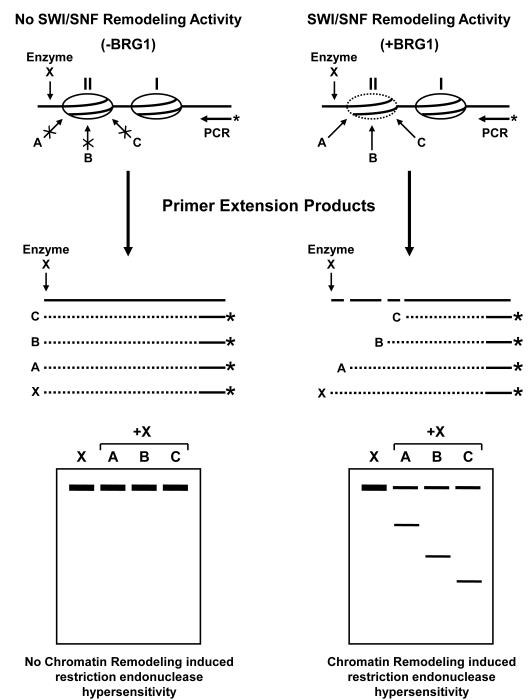
Figure 1.
Schematic of the restriction enzyme hypersensitivity assay used to evaluate chromatin structure and detect SWI/SNF-mediated chromatin remodeling activity. Nuclei are isolated from naïve or hormone-treated cells with (+BRG1) or without (-BRG1) expression of a functional SWI/SNF chromatin remodeling complex. The chromatin is partially digested with various restriction endonucleases (A, B, or C) that target the predicted hypersensitive region (II). After purification, the genomic DNA is digested to competition with a second restriction enzyme which cut outside the hypersensitive region (enzyme X). The resulting DNA is purified and analyzed by reiterative primer extension using Taq polymerase and a 32P-labeled oligonucleotide specific for the target of interest. The amplified products are resolved on a denaturing polyacrylamide gel and exposed to a PhosphorImager screen to detect the presence of bands that correspond, in length, to the predicted in vivo restriction endonuclease cleavage pattern. Re-digestion with the second restriction enzyme (in vitro digest) serves as an internal standard to ensure equal amounts of template DNA was loaded in each reaction. The prediction in this model would suggest that the SWI/SNF complex targets regions within nucleosome II where it remodeling the chromatin structure to allow access to transcription factors or, in this case, restriction endonucleases to the genomic DNA. In the absences of BRG1, SWI/SNF is unable to remodel nucleosome II where it act to block restriction enzyme access to the target DNA.