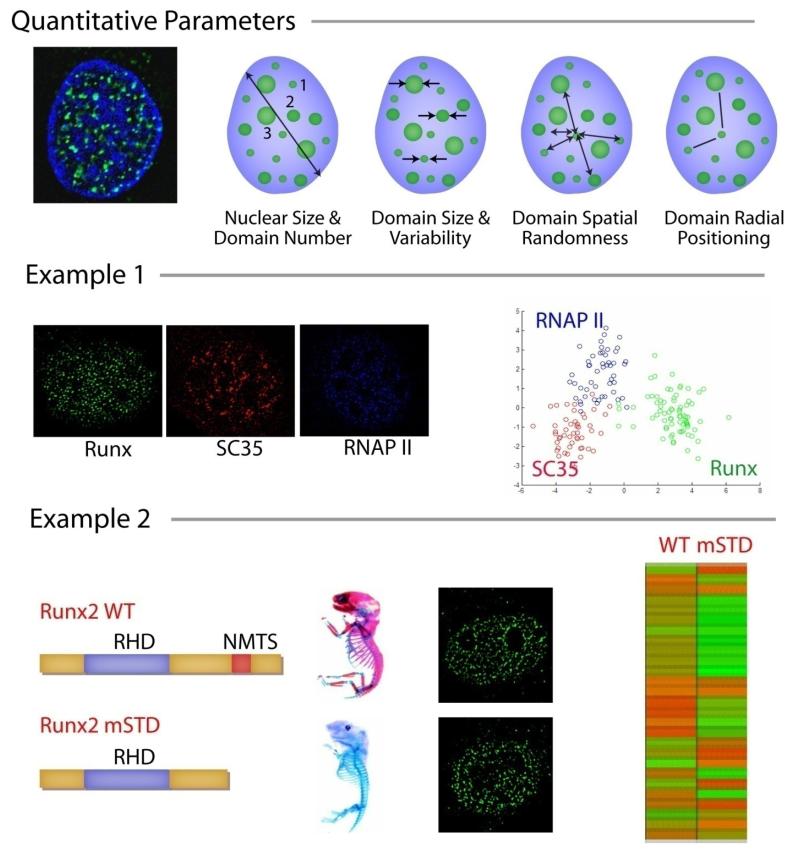
Figure 4. Intranuclear Informatics.
This figure shows how intranuclear informatics can be used to examine nuclear alterations in cancer cells compared with normal cells. The top panel shows the conceptual framework for the quantitation of subnuclear organization by intranuclear informatics. Four main groups of parameters, selected based on inherent biological variability, can be examined. Example 1. Regulatory proteins with different activities can be subjected to Intranuclear Informatics analysis, which assigns each protein a unique architectural signature. The overlap between the architectural signatures of different proteins is often correlated to their functional overlap. Shown here are Runx transcription factor, SC35 splicing protein, and RNA Pol II. Example 2. The subnuclear organization of Runx domains is linked with subnuclear targeting, biological function, and disease. Biologically active Runx2 and inactive subnuclear targeting defective mutant (mSTD) show distinct architectural signatures, indicating that the biological activity of a protein can be defined and quantified as subnuclear organization.