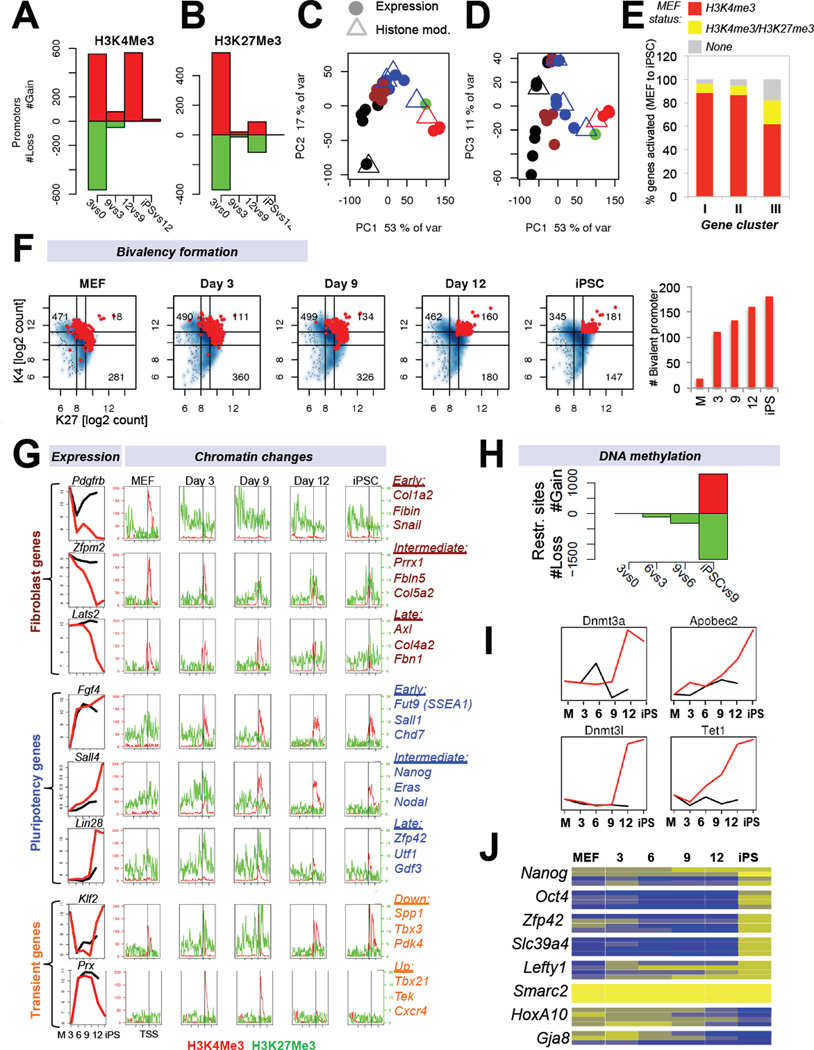
Figure 5. Histone and DNA methylation dynamics during cellular reprogramming (see also Figure S4).
(A, B) Enrichment for H3K4me3 or H3K27me3 at promoters of differentially expressed genes in progressing intermediates. (C, D) Superimposition of principal component analyses for genes enriched in H3K4me3 (C) or H3K27me3 (D)(triangles) with gene expression data (circles) of the same cell populations (see Figure 2A for color coding). (E) Display of activated genes from gene expression categories I, II and III (see Figure 2E) in relation to their chromatin status in MEFs. (F) Number of differentially expressed genes that become bivalent (H3K27me3 and H3K4me3 enriched) during reprogramming (red dots = bivalent promoters) and quantification. (G) Integration of gene expression and histone modifications data defines subsets of genes with characteristic expression changes. Shown are examples of fibroblast-associated (top), pluripotency-associated (center) and transiently changing genes (bottom). (H) Number of genes, which change DNA methylation status in progressing cell populations during reprogramming as determined by genome-wide methylation analysis. (I) Expression dynamics of candidate genes associated with DNA methylation and demethylation. (J) Heatmap of DNA methylation analysis of specific CpGs (boxes) in the promoter regions of indicated genes during reprogramming using EpiTYPER DNA methylation analyses. Yellow indicates 0% methylation and blue represents 100% methylation.