Abstract
The product of p53-induced gene 1 is a member of the galectin family, i.e., galectin-7 (Gal-7). To move beyond structural data by X-ray diffraction, we initiated the study of the lectin by nuclear magnetic resonance (NMR) and circular dichroism spectroscopies, and molecular dynamics (MD) simulations. In concert, our results indicate that lactose binding to human Gal-7 induces long-range effects (minor conformational shifts and changes in structural dynamics) throughout the protein that result in stabilization of the dimer state, with evidence for positive cooperativity. Monte Carlo fits of 15N-Gal-7 HSQC titrations with lactose using a two-site model yield K1 = 0.9 ± 0.6 × 103 M−1 and K2 = 3.4 ± 0.8 × 103 M−1. Ligand binding-induced stabilization of the Gal-7 dimer was supported by several lines of evidence: MD-based calculations of interaction energies between ligand-loaded and ligand-free states, gel filtration data and hetero-FRET spectroscopy that indicate a highly reduced tendency for dimer dissociation in the presence of lactose, CD-based thermal denaturation showing that the transition temperature of the lectin is significantly increased in the presence of lactose, and saturation transfer difference (STD) NMR using a molecular probe of the monomer state whose presence is diminished in the presence of lactose. MD simulations with the half-loaded ligand-bound state also provided insight into how allosteric signaling may occur. Overall, our results reveal long-range effects on Gal-7 structure and dynamics, which factor into entropic contributions to ligand binding and allow further comparisons with other members of the galectin family.
Keywords: circular dichroism, FRET, galectin, molecular dynamics, NMR
Introduction
Comparative structural analysis of members of a protein family is a means of tracing the impact of sequence variations seen in phylogenesis and assigning functionality to them. The emerging broad range of effector activities of adhesion/growth-regulatory galectins gives ample incentive to focus on their systematic study (Cooper 2002; Villalobo et al. 2006; Gabius et al. 2011). Toward this end, we have initiated here the structural analysis of human galectin-7 (Gal-7) in solution by nuclear magnetic resonance (NMR) spectroscopy. Initially, Gal-7 was identified as human keratinocyte protein down-regulated in SV40-transformed cells (Madsen et al. 1995) moderately repressed by retinoic acid (Magnaldo et al. 1995). It has attracted special attention since its gene was referred to as p53-induced gene 1, which was based on its marked activity increase upon p53 expression prior to the onset of apoptosis in human D.L. Dexter-1 (DLD-1) colon carcinoma cells (Polyak et al. 1997). Fittingly, Gal-7 appears to be involved in the process of ultraviolet B (UVB, 280-315 nm)-elicited apoptosis in keratinocytes (Bernerd et al. 1999). Beyond in vitro studies, the value of lectin has already been proved as a diagnostic marker in differential tumor diagnosis and as an indicator of prognostic evaluations with the aim to optimize postsurgical treatment in head and neck tumors by stratification (Saussez et al. 2006; Remmelink et al. 2011).
Mechanistically, Gal-7 is capable of triggering growth regulation via intracellular protein targets, such as its interaction with bcl-2 or carbohydrate-dependent binding to cell surface glycans that are then cross-linked (Kuwabara et al. 2002; Kopitz et al. 2003; Sturm et al. 2004; Villeneuve et al. 2011). The latter property was described to be sensitive to the degree of N-glycan branching and its interaction with the pan-galectin ligand N-acetyllactosamine encompassing a fairly large entropic penalty as revealed by calorimetric analysis (Ahmad et al. 2002; Hirabayashi et al. 2002; Dam et al. 2005). Dissecting contributions to the entropic term cannot be readily done on the basis of the crystal structure. Of note, crystal packing precluded access of a carbohydrate ligand to one subunit in the homodimer, which has a rather large interface of 1484 Å2 (compared with 1093 Å2 in the canonical galectin-1 (Gal-1 dimer)) (Leonidas et al. 1998). This size of the interface underlies the homodimer status in solution, as measured by gel filtration, mass spectrometry and ultracentrifugation, also inferred by hemagglutination and reactivity to glycoclusters (André et al. 2001, 2004; Kopitz et al. 2003; Morris et al. 2004).
Having recently reported resonance assignments for human Gal-7 (Nesmelova et al. 2012), it was now possible to assess the effect of ligand binding on the protein, with an eye on structural long-range consequences by ligand loading. By combining NMR experiments using uniformly 15N-labeled Gal-7, circular dichroism (CD) studies and molecular dynamics (MD) simulations, we herein provide insight into structural responses of the lectin to ligand association. Moreover, these data provide the opportunity for comparisons with the respective behaviors of Gal-1 and -3, as previously reported (Umemoto et al. 2003; Diehl et al. 2009, 2010; Nesmelova et al. 2010).
Results
Lactose binding
Heteronuclear Single Quantum Coherence (HSQC) spectra of 15N-enriched Gal-7 (15N-Gal-7) are shown in Figure 1 as a function of lactose concentration. While some resonances are relatively highly chemically shifted during the titration, others are less shifted, if at all. Net chemical shift changes, Δδ, for all amino acid residues are shown in Figure 2A, and residues showing the greatest changes are highlighted in color on the a subunit from the lactose-loaded dimer structure in Figure 2B. Not unexpectedly, many of the most highly shifted resonances (red and orange) belong to residues within several Å of the bound lactose molecule, with the magnitude of Δδ generally decreasing as the distance from the lactose-binding site increases. Nonetheless, some significantly perturbed residues (pink) are located at sites more distant from the ligand-binding region, in particular on the backside of the β-sandwich at the dimer interface (Figure 2B, right). In this regard, carbohydrate ligand binding induces long-range conformational and/or dynamical effects in the lectin.
Fig. 1.
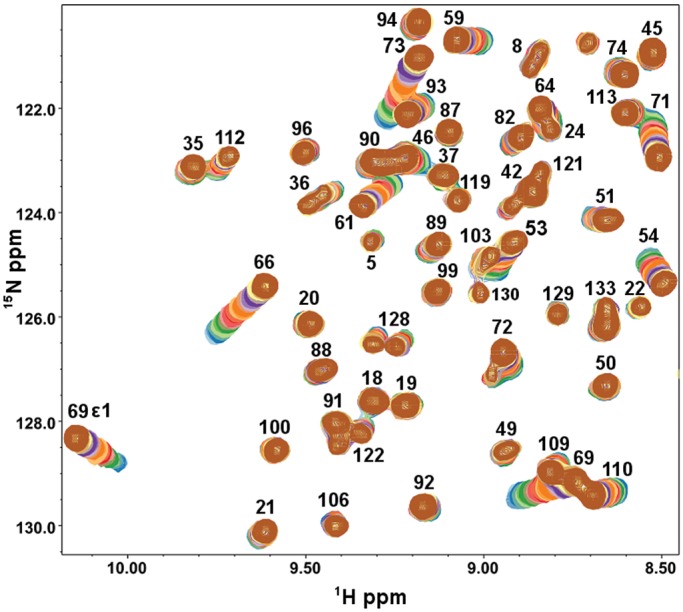
Expansions from 15N–1H HSQC spectra of Gal-7 are shown, with 23 overlays, one for Gal-7 alone (blue cross-peaks) and the others for Gal-7 in the presence of lactose from 0.01 to 45 mM (brown). Gal-7 was dissolved in an aqueous (95% 1H2O/5% 2H2O) solution of 20 mM potassium phosphate buffer, pH 7, containing 2 mM DTT, at 30°C.
Fig. 2.
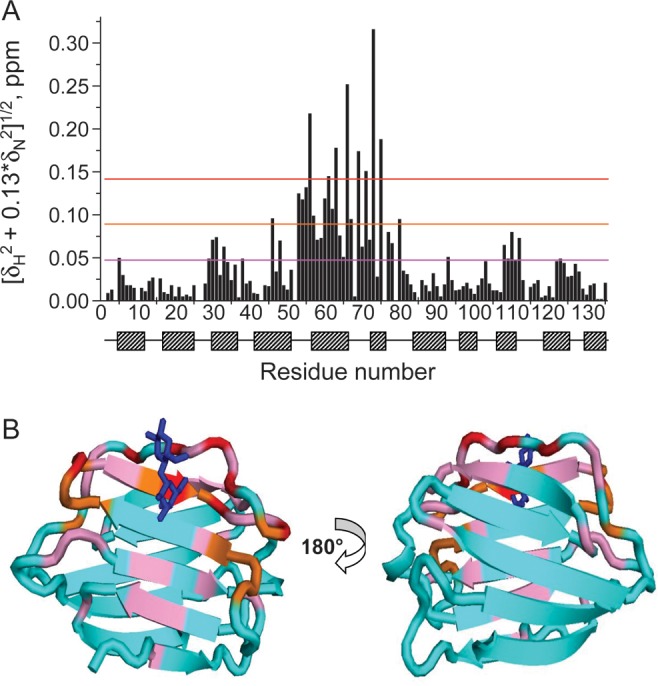
(A) 1H–15N-weighted chemical shift differences (Δδ) for backbone NH groups in lactose-free and lactose-loaded Gal-7 monomer subunits are shown, along with the positions of β-strands and loops/turns in Gal-7. (B) Structure of the lactose-loaded Gal-7 dimer is shown, with Δδ values from panel (A) highlighted in red for the most shifted resonances, followed by orange, pink, and then cyan for those resonances that are minimally or not at all shifted.
To obtain association equilibrium constants for lactose binding to Gal-7, we curve fit these HSQC titration data using one- and two-site binding models and the Monte Carlo procedure described previously (Nesmelova et al. 2010; Miller et al. 2011). Using average values for the six most highly chemically shifted resonances, the Hill plot in Figure 3A shows that the one-site model fit (K1 = K2, red circles) deviates considerably from the actual data points (open squares), whereas the two-site model fit (black circles) represents these data relatively accurately. Figure 3B shows individual titration curves for the same six resonances, with lines showing fits using the two-site model. Figure 3C plots K1 and K2 values derived from two-site model fits for the twenty most highly chemically shifted resonances, considering 1H and 15N resonances individually. Each point represents one K1/K2 pair and the straight diagonal line indicates K1 = K2. All points fall on that side of the diagonal where K1 < K2. The averages of these values are K1 = 0.9 ± 0.6 × 103 M−1 and K2 = 3.4 ± 0.8 × 103M−1. These data indicate that lactose binding to Gal-7 occurs with positive cooperativity (Hill coefficient n = 1.32) such that ligand binding at the first site should induce conformational and/or dynamic changes through the protein at the other site to promote greater ligand binding affinity.
Fig. 3.
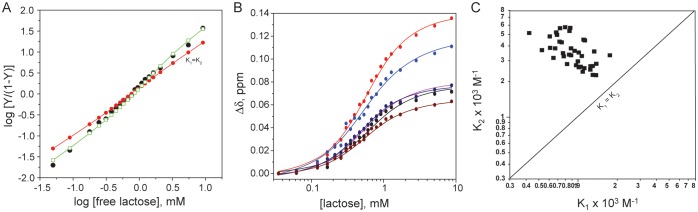
(A) Hill plot showing the average of the six most chemically shifted resonances plotted as log fraction bound/(1-fraction bound) vs. log free ligand concentration. Actual HSQC data points are shown as green squares, while fits with the one-site and two-site model are shown as filled red circles and filled black circles, respectively, as discussed in the text. (B) Lactose titration showing 15N–1H-weighted chemical shifts for resonances of some residues vs. the concentration of lactose. Optimal curve fitting using a two-site model (solid lines) gives values for the equilibrium binding constants of K1 = 0.9 ± 0.6 × 103 M−1 and K2 = 3.4 ± 0.8 × 103 M−1. (C) Plot of actual K1 and K2 values resulting from Monte Carlo fits to 20 titration curves showing the greatest chemical shift differences (1H and 15N chemical shifts treated separately), indicating that ligand binding may occur with positive cooperativity as discussed in the text.
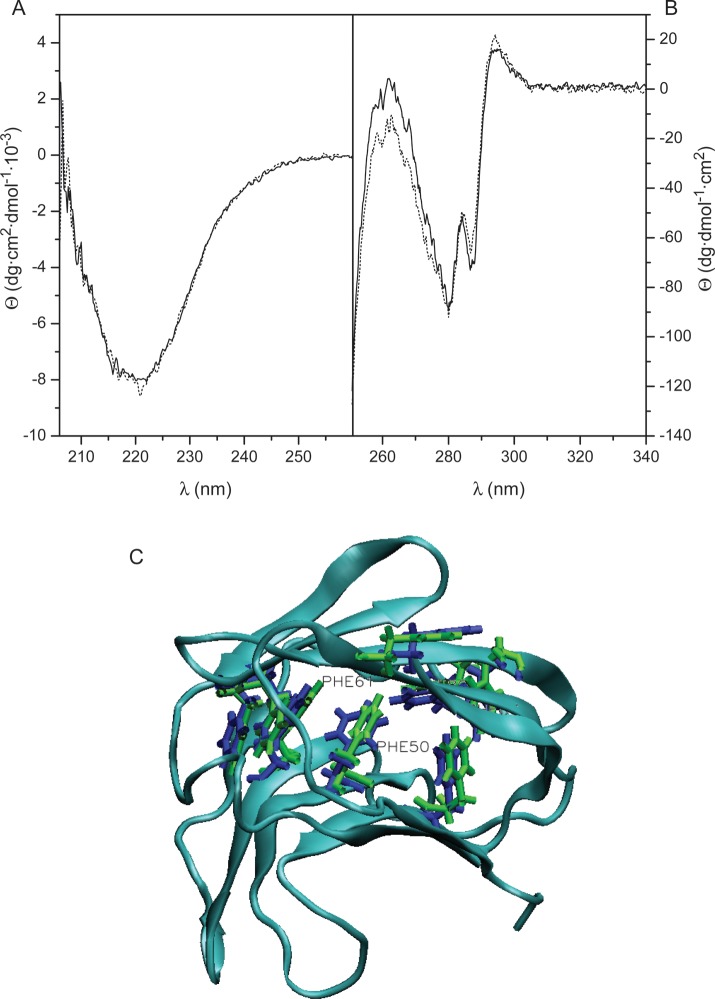
On the crystallographic level, structures of ligand-free (protein data bank (PDB) 1BKZ) and fully galactose-loaded (PDB 2GAL) Gal-7 superimpose with an a Root-Mean-Square Deviation (RMSD) over all Cα atoms of 0.66 Å, and structures of ligand-free and Gal-7 with one bound lactose molecule (PDB 4GAL) superimpose with an RMSD over all Cα atoms of 0.26 Å. In addition, RMSD over backbone atoms (all but the first four N-terminal residues) of the two subunits in the half-loaded lactose-bound state is 0.54 Å, with a total RMSD for heavy atoms of 0.98 Å. These X-ray data indicate that there are no major conformational differences in the crystal structures between ligand-bound and ligand-free Gal-7, or for that matter between/among other galectins studied at this level (Nesmelova et al. 2008a). This conclusion is echoed by our NMR data (e.g., Figure 1), where the Gal-7 HSQC spectral fingerprint (i.e., resonance dispersion) is very similar between lactose-bound and free states, indicating that lactose binding does not induce major conformational changes in the lectin. In addition, far-UV CD spectroscopy does not reveal any apparent differences in secondary structural features of Gal-7 in the absence and presence of lactose (Figure 4A). However, the near-UV CD spectrum (Figure 4B) does reflect some change at tertiary-structure level. This was characterized by the presence of a broad negative region with well-defined bands centered at 280 and 287 nm, in the tyrosine/tryptophan region, and a positive signal at 294 nm, attributable to the single Gal-7 tryptophan residue, located at the carbohydrate-binding site. In the presence of lactose, small but unequivocally consistent changes in ellipticity at 287 (decrease in negative ellipticity) and 294 nm (increase in positive ellipticity) were observed, independently of the protein concentration and temperature used for acquisition of the spectra. These changes are compatible with a variation in the contribution of positive 1La and negative 1Lb transitions of the Trp residue, reflecting a modification of its environment upon ligand binding due most likely to the alignment (stacking) of the indole ring with a lactose molecule. In addition, there is an increase in negative ellipticity at 257–267 nm. Considering that the described changes in Trp transitions should contribute with positive ellipticity to this spectral region, the appearance of a trough at 262 nm can be interpreted as a change in the chiroptical properties of phenylalanine residues, suggesting that the hydrophobic core of the β-sandwich fold, in which 7 of the 8 phenylalanine residues in Gal-7 are accommodated, is sensitive to binding of the ligand. Nevertheless, these minimal near-UV CD spectral changes point to the absence of major ligand-induced conformational changes in the protein, consistent with our NMR results.
Fig. 4.
Representative CD spectra obtained for Gal-7. Far- (A) and near-UV CD spectra (B) were obtained at 30°C for 0.2 mg/mL (13 μM monomer) and 1 mg/mL (67 μM monomer) solutions, respectively, of Gal-7 in 20 mM potassium phosphate buffer, pH 7.0, containing 2 mM DTT, in the absence (solid lines) and presence of 10 mM lactose (dotted lines). (C) MD simulation-derived average structures of ligand-free (PDB 1BKZ) and galactose-loaded (PDB 2GAL). Gal-7 subunits are shown with Phe side chains from ligand-free and galactose-bound Gal-7 highlighted in blue and green, respectively. The backbone ribbon shown is an average of both free and bound structures.
In addition to subtle conformational shifts, ligand binding-induced effects in Gal-7 may be explained by changes in internal motions or conformational fluctuations. MD simulations are well suited to provide insight into these changes, and have the added advantage of being able to investigate the half-loaded ligand-bound state. For these reasons, we performed MD simulations on the Gal-7 dimer and analyzed root mean-square fluctuations (RMSF) of backbone and side-chain atoms for three states of the protein: the ligand-free state (PDB 1BKZ), the half-loaded lactose-bound state (PDB 4GAL) and the galactose-loaded state (PDB 2GAL) “in lieu” of the fully lactose-loaded state which has not been determined. RMSF values reflect changes in motional amplitudes such that a smaller value indicates reduced internal motions and vice versa.
These results are consistent with near-UV CD data in that average positions of most Phe side chains appear to be minimally affected by ligand binding (Figure 4C). Two exceptions are F50 and F61, whose phenyl rings are re-oriented upon ligand binding. Moreover, ligand-induced changes in RMSF values for Phe side chains (Table I) indicate that internal motions are generally significantly reduced in ligand-loaded (galactose and lactose) states vis-à-vis ligand-free Gal-7. The average of side-chain RMSF values is 0.96 for ligand-free Gal-7 and 0.79 for fully galactose-loaded Gal-7. We also find that Phe side-chain motions in both subunits of the half-loaded state are similarly reduced (on average, 0.66 and 0.68, respectively) vis-à-vis ligand-free Gal-7. Changes in Phe side-chain motion could account for the increased negative ellipticity observed in the Phe region of near-UV CD spectra and, most importantly, suggest that binding of the first ligand induces an allosteric transition in the ligand-free subunit, making the protein “terroir” more like that in the subunit with ligand and preparing the second binding site for hosting the ligand more readily, a view that is consistent with our observation of positive cooperativity in ligand binding.
Table I.
RMSF of backbone Cα atoms and RMSF for side chains of Phe residues of Gal-7 in loaded, ligand-free and half-loaded states
| Residual no. | Backbone |
Side chains |
||||||
|---|---|---|---|---|---|---|---|---|
| Gal-7 free | Loaded with 2 galactose molecules | Gal-7 loaded with 1 lactose molecule |
Ligand-free Gal-7 | Loaded with 2 galactose molecules | Gal-7 loaded with 1 lactose molecule |
|||
| Ligand-free subunit | Loaded subunit | Ligand-free subunit | Loaded subunit | |||||
| 32 | 0.76 | 0.58 | 0.51 | 0.53 | 0.92 ± 0.15 | 0.74 ± 0.15 | 0.66 ± 0.14 | 0.77 ± 0.22 |
| 50 | 0.51 | 0.45 | 0.44 | 0.41 | 0.76 ± 0.22 | 0.91 ± 0.4 | 0.71 ± 0.27 | 0.58 ± 0.14 |
| 61 | 0.66 | 0.56 | 0.5 | 0.44 | 0.81 ± 0.1 | 0.72 ± 0.12 | 0.64 ± 0.11 | 0.56 ± 0.1 |
| 80 | 1.03 | 0.9 | 0.62 | 0.77 | 1.0 ± 0.14 | 0.91 ± 0.13 | 0.68 ± 0.08 | 0.83 ± 0.08 |
| 86 | 1.2 | 0.74 | 0.55 | 0.53 | 1.2 ± 0.09 | 0.81 ± 0.09 | 0.64 ± 0.08 | 0.76 ± 0.22 |
| 97 | 0.73 | 0.55 | 0.49 | 0.42 | 0.86 ± 0.12 | 0.7 ± 0.12 | 0.73 ± 0.21 | 0.57 ± 0.12 |
| 109 | 0.82 | 0.65 | 0.54 | 0.44 | 1.04 ± 0.1 | 0.75 ± 0.1 | 0.75 ± 0.17 | 0.54 ± 0.08 |
| 135 | 0.89 | 0.58 | 0.51 | 0.5 | 1.11 ± 0.27 | 0.79 ± 0.18 | 0.65 ± 0.12 | 0.66 ± 0.13 |
Standard deviations are shown as ± values from average values.
For ligand-free Gal-7 and Gal-7 loaded with two galactose molecules, averages of both subunits are shown.
Additional ligand binding-induced changes in internal motions
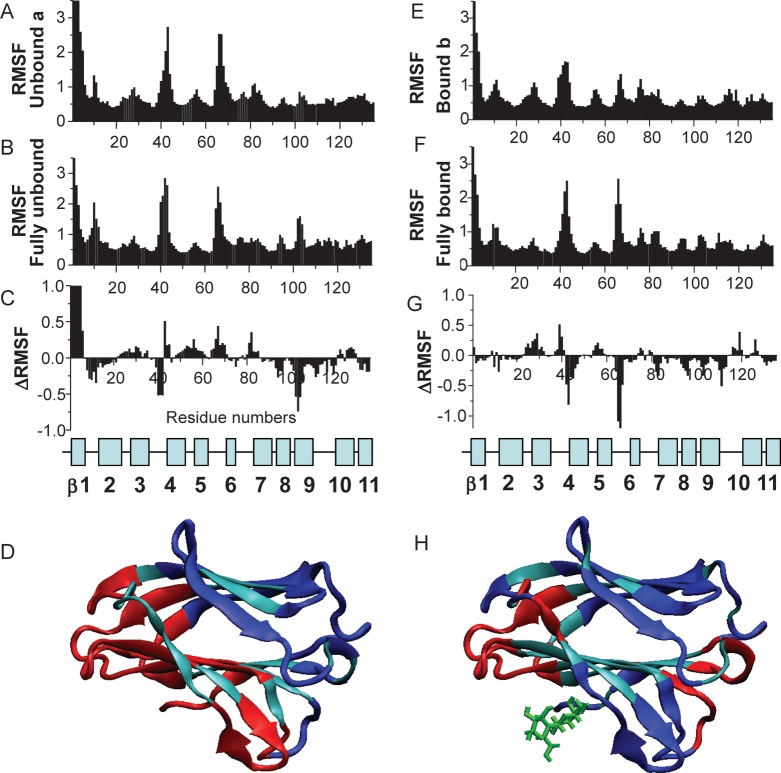
Figure 5A–D shows RMSF values for Cα atoms of the ligand-free subunit from the half-loaded state (A), the average of the two subunits from fully ligand-free state (B) and the difference between the two (C = A − B). Since dynamics for backbone NH atoms follow essentially the same trends, these data are not shown. Both structures display similar RMSF distributions through the sequence, with N-terminal residues and inter-strand loops (especially loops connecting strands β3-β4, β5-β6 and β8-β9) showing the largest conformational fluctuations and C-terminal motions being restricted by interactions with residues from the second subunit. Of greater interest here is the difference between these two distributions (Figure 5C), which is highlighted on the Gal-7 subunit in Figure 5D. The blue and red colors indicate a decrease or an increase in RMSF values, respectively, while the aqua color indicates minimal change, if any. Of note, motions of residues within and around the lactose-binding site generally become more mobile (red) in the ligand-free subunit, when lactose is bound to the other subunit (half-loaded state). In contrast, those on the opposite face of the β-sandwich (that includes residues at the dimer interface) generally become more motionally restricted (blue), similar to Phe residues within the hydrophobic core, as discussed above. The increase in mobility of residues at the ligand-free site itself may facilitate docking or induced fit of the second ligand, with the expected entropic penalty to binding being compensated for by decreased internal motions elsewhere in the protein.
Fig. 5.
RMSF values for Cα atoms are shown for ligand-free subunit a, when one lactose moiety is associated with subunit b (A); for the average of completely ligand-free Gal-7 (B) and for the difference A minus B (C). The position of β-strands and loops in Gal-7 is shown below. (D) The changes of RMSF values are highlighted for subunit a of the half-loaded Gal-7 state. The blue and red colors indicate decreasing and increasing RMSF values, respectively, as a result of ligand binding. Unaffected residues are shown in cyan. RMSF values for Cα atoms are shown for the ligand-loaded subunit b in the half-loaded state (E); for the average of fully loaded (galactose) Gal-7 (F) and for the difference E minus F (G). The position of β-strands and loops in Gal-7 is shown below. (H) The changes of RMSF values are highlighted for subunit b of the half-loaded Gal-7 state. The blue and red colors indicate decreasing and increasing RMSF values, respectively.
Figure 5E–H shows RMSF values for Cα atoms of the ligand-loaded subunit in the half-loaded state (E), the average of the two subunits in the fully galactose-loaded state (F) and the difference between the two (G = E − F). The difference plot (Figure 5G) indicates that internal motions in the ligand-loaded subunit of half-loaded lectin are overall more restricted than are those in the fully loaded dimer. Segments showing the greatest differences are highlighted in blue and red on the lactose-loaded subunit of the dimer (Figure 5H). In this comparison, motional differences between the two faces of the β-sandwich are not as distinct as in the ligand-free subunit (Figure 5D). In particular, changes in internal motions of residues at the lactose-binding site are mixed, with residues on one side of the binding site showing decreased mobility and others in surrounding segments showing less restricted mobility. Residues at the dimer interface generally become more motionally restricted upon lactose binding. Overall, however, side-chain motions are on average increased upon ligand binding (data not shown). The mean RMSF value for side chains of ligand-free lectin is 1.22, whereas it is 1.37 for galactose-loaded Gal-7. The extent of these changes in terms of conformational entropy will factor into the thermodynamics of ligand binding and may also affect the architecture of the dimer interface to influence dimer stability.
Ligand binding enhances dimer stability
Using the molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) method (Kollman et al. 2000) and MD simulations, we determined van der Waals (EVdW), electrostatic (EPB) and nonpolar solvation (ESASA) contributions to inter-subunit interaction energies (Nesmelova et al. 2008b). Lactose binding to Gal-7 induces significant changes to inter-residue interaction energies, not surprisingly, between residues at the ligand-binding site (Table II), but also with equal and even greater magnitude between other residues within and between subunits. Inter-subunit residues that contribute most favorably to the energetics of dimerization are R14, R20, R22, E87, L89, D94, D95, D103, A104, Q105, D130 and F135, as highlighted in the dimer structure shown in Figure 6. Although EVdW, EPB and ESASA all contribute favorably to the monomer–monomer interaction (Table III), the largest stabilizing effect derived from ligand binding appears to come from electrostatic contributions, likely via formation of salt bridges, e.g., R20-E87. For ligand-loaded and ligand-free dimers, average interaction energies are −107 kcal/mol with two ligands (galactose) bound, −98 kcal/mol for one ligand (lactose) bound and −88.2 kcal/mol for the completely ligand-free state, with differences in energies, ΔΔE, from the ligand-free state being −10 kcal/mol and −19 kcal/mol, respectively. The favorable energy change upon binding the first ligand is followed by an additionally equally favorable energy change upon binding the second ligand.
Table II.
Energetic contributions (kcal/mol) to lactose loading for individual amino acids at the binding site
| Residue | EPB | EVdW | Total |
|---|---|---|---|
| R31 | 0.13 | −0.28 | −0.15 |
| H33 | 0 | −0.19 | −0.19 |
| H49 | −0.9 | −2.14 | −3.04 |
| N51 | 0.1 | −1.31 | −1.21 |
| R53 | −2.1 | −0.34 | −2.44 |
| E58 | 0.14 | −0.28 | −0.14 |
| V60 | 0 | −1.5 | −1.5 |
| N62 | 0 | −1.62 | −1.62 |
| K64 | 0.22 | −0.27 | −0.05 |
| W69 | −0.34 | −5.85 | −6.19 |
| R71 | −0.12 | −0.12 | −0.24 |
| E72 | 0.12 | −0.6 | −0.48 |
| E73 | −0.14 | −0.1 | −0.24 |
| R74 | −0.12 | −0.47 | −0.59 |
Fig. 6.
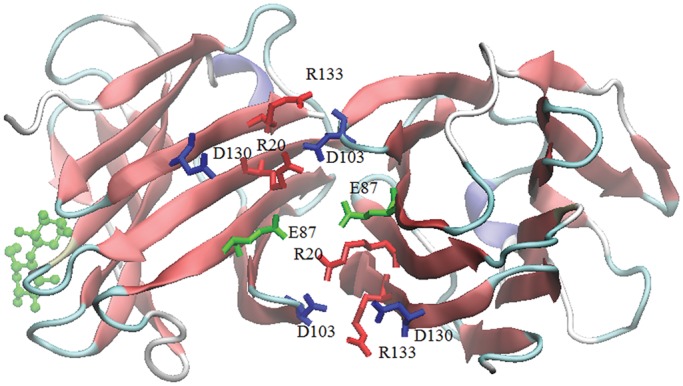
The Gal-7 dimer with one lactose bound is shown, and residues involved in the dimer interface are indicated and colored according to the type of residue. Arginines are in red, glutamates are in green and aspartates are in blue.
Table III.
Interaction energies (kcal/mol) among residues at the Gal-7 dimer interface
| Dimer | PB | VDW | SASA | Total |
|---|---|---|---|---|
| Ligand-free Gal-7 | −34.3 ± 5.1 | −45.7 ± 7.0 | −8.2 ± 0.5 | −88.2 |
| Half-loaded lactose-bound Gal-7 | −43.6 ± 5.0 | −46.1 ± 6.0 | −8.3 ± 0.3 | −98.0 |
| Fully loaded galactose-bound Gal-7 | −50.4 ± 5.4 | −47.5 ± 8.5 | −9.1 ± 0.5 | −107 |
This theoretical increase in ligand binding-induced dimer stability was validated experimentally in several assays: Gel filtration fast protein liquid chromatography (FPLC), CD-based thermal denaturation studies, Fluorescence Resonance Energy Transfer (FRET) using Edans- and Dabcyl-labeled Gal-7 and STD NMR with a molecular probe to Gal-7 monomers. Previous analyses of quaternary structure using nanoESI MS, ultracentrifugation and gel filtration showed that Gal-7 is predominantly dimeric at concentrations around 0.15 mg/mL (10 μM in monomer Gal-7) and above (Kopitz et al. 2003; André et al. 2004; Morris et al. 2004). Here, we extended these studies and examined the behavior in gel filtration chromatography of increasingly diluted solutions of Gal-7 (from 2.4 to 0.04 mg/mL or 160 to 2.6 μM monomer Gal-7) resulting in eluted concentrations of 0.3–0.005 mg/mL (20–0.3 μM monomer Gal-7), by monitoring the protein elution at 215 nm. Our results show that in the absence of lactose, the elution time from the FPLC column is progressively increased as the concentration of eluted lectin is decreased (Figure 7A, open squares). Elution times for protein markers carbonic anhydrase (29 kDa) and cytochrome c (12.4 kDa) were 25.7 ± 0.5 and 29.4 ± 0.5 min, respectively. Relative to these elution markers, we find that Gal-7 elutes mostly as a dimer at the highest concentrations, in agreement with previous findings, and mostly as a monomer at the lowest concentrations under these conditions, consistent with dimer dissociation over the protein concentration range investigated. The mid-point for the dimer dissociation curve is about 0.025 mg/mL (1.6 μM), yielding an apparent equilibrium dissociation constant, Kd, of ∼1.7 × 10−6 M. Lactose binding shifts this Gal-7 dimer Kd value in favor of the dimer state. In the presence of 10 mM lactose (Figure 7A, solid squares), dimers in FPLC show no apparent dissociation over the concentration range investigated, indicating that the Kd value in the lactose-loaded state should be much less than the lowest concentration used, i.e., <<0.4 × 10−6 M (at least 30-fold smaller considering the mass action law). In comparison, no shift in the elution position of Gal-1 was observed within the same range of concentrations, either in the absence and presence of lactose (data not shown), indicating that the Kd for Gal-1 (ligand-loaded or ligand-free) is much lower than that for Gal-7.
Fig. 7.
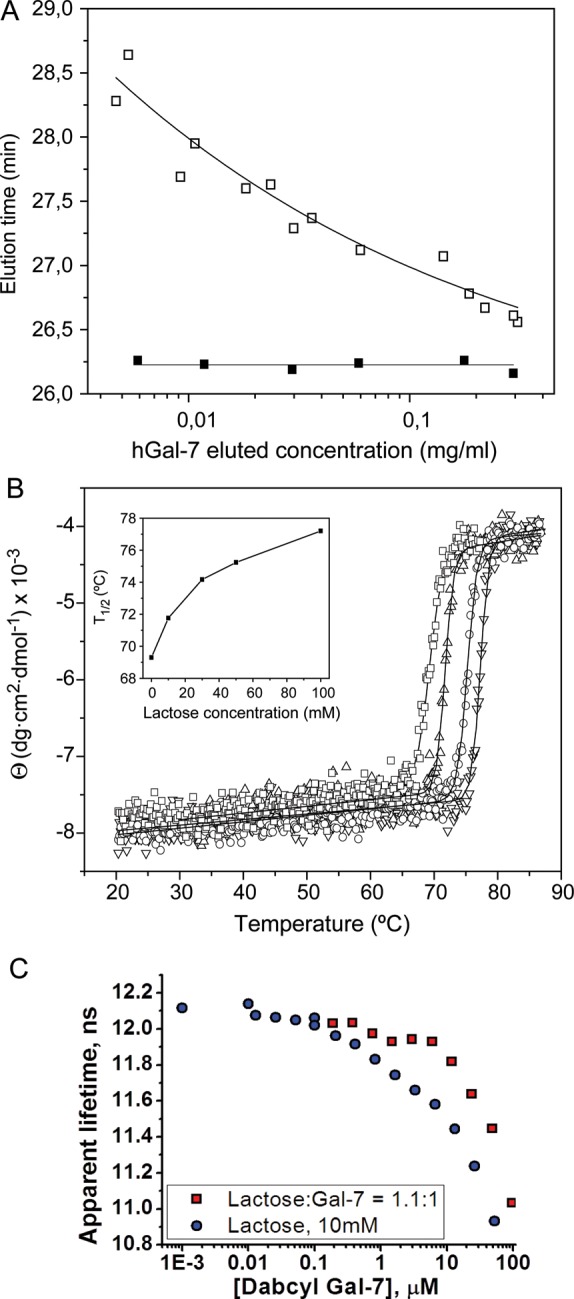
(A) Variation with protein concentration of the elution time of Gal-7 in gel filtration FPLC. 100 µL samples of Gal-7 at 0.04–2.4 mg/mL (2.6–160 μM monomer) were chromatographed in a Superose 12 column equilibrated at 30°C with 20 mM potassium phosphate buffer, pH 7.0, containing 2 mM DTT and 0.02% NaN3, in the absence (open squares) or presence of 10 mM lactose (filled squares). The concentration of the eluted protein was calculated using the dilution factor estimated for each peak from the ratio of width at half-height of the peak to the injection volume. (B) Effect of lactose on the stability of Gal-7 against thermal denaturation. The ellipticity at 220 nm of Gal-7 was monitored as a function of temperature, in the absence (open squares) and presence of 10 mM (open triangle), 50 mM (open circles) and 0.1 M (inverted triangle) lactose. The continuous lines correspond to the fit of a sigmoidal function to experimental data. Inset: Variation of the T1/2 with increasing lactose concentrations. (C) Hetero-FRET data showing the dependence of the apparent lifetime of donor fluorescence (fixed concentration of Edans-labeled Gal-7) as a function of the concentration of acceptor DABCYL-labeled Gal-7. The curve with red data points shows a molar ratio of lactose:Gal-7 of 1.1:1, and the blue data points are for lactose at 10 mM.
Ligand binding-induced stabilization of Gal-7 structure (tertiary and quarternary) was supported by CD-based thermal denaturation studies (Figure 7B). Thermal denaturation of Gal-7 resulted in irreversible loss of secondary structure with subsequent precipitation of the protein. The process was monitored by measuring the decrease in negative ellipticity at 220 nm with temperature. A phenomenological analysis of the experimental curves using a Gaussian/sigmoidal function yielded a value for T1/2 of 69.3 ± 0.1°C. This T1/2 is very close to that previously determined for Gal-1 (71.1 ± 0.2°C), although the denaturation process for this protein was not followed by precipitation and the observed temperature-induced changes were partially reversible (Solís et al. 2010). The presence of 10 mM lactose, a concentration producing a substantial stabilization of the Gal-7 dimer as evidenced by FPLC, also induced an increase of 2.4 ± 0.03°C in the T1/2 value. The thermal stability of Gal-7 was further progressively enhanced in the presence of increasing concentrations of the sugar (Figure 7B), reaching a T1/2 of 77.2 ± 0.1°C at 0.1 M lactose. This behavior, which is similar to that exhibited by Gal-1 (T1/2 increasing for this protein by 7.4°C in the presence of 0.1 M lactose (Solís et al. 2010)), is consistent with the ligand being bound by the two galectins in the folded form. Thus, the different dimerization modes of Gal-7 and -1 are not reflected in major differences in thermal stability or in the effect of ligand binding on the stabilization of the proteins against thermal denaturation, i.e., both galectin dimers are thermodynamically stabilized by ligand binding.
Next, we thio-labeled Gal-7 at its sole cysteine (C38) with two fluorophores (Edans and DABCYL) and used hetero-FRET to assess the Gal-7 monomer–dimer equilibrium. Figure 7C shows the dependence of the apparent lifetime of donor fluorescence (fixed concentration of Edans-labeled Gal-7) as a function of the concentration of acceptor Dabcyl-labeled Gal-7. We performed the hetero-FRET experiment in the presence of 10 mM lactose and in the presence of a low concentration of lactose (1.1:1 molar ratio) to maintain galectin fold stability. Figure 7C shows that the mid-points in these curves suggest that the dimer equilibrium dissociation constants are ∼8 and 30 μM, respectively, which are considerably higher than those estimated from FPLC. This is likely the result of limited labeling (∼17%) and label-induced changes in the conformation and fold stability in fluorophore-labeled Gal-7. Nevertheless, when Gal-7 is saturated with lactose ligand, the dimer state is stabilized, consistent with our other data.
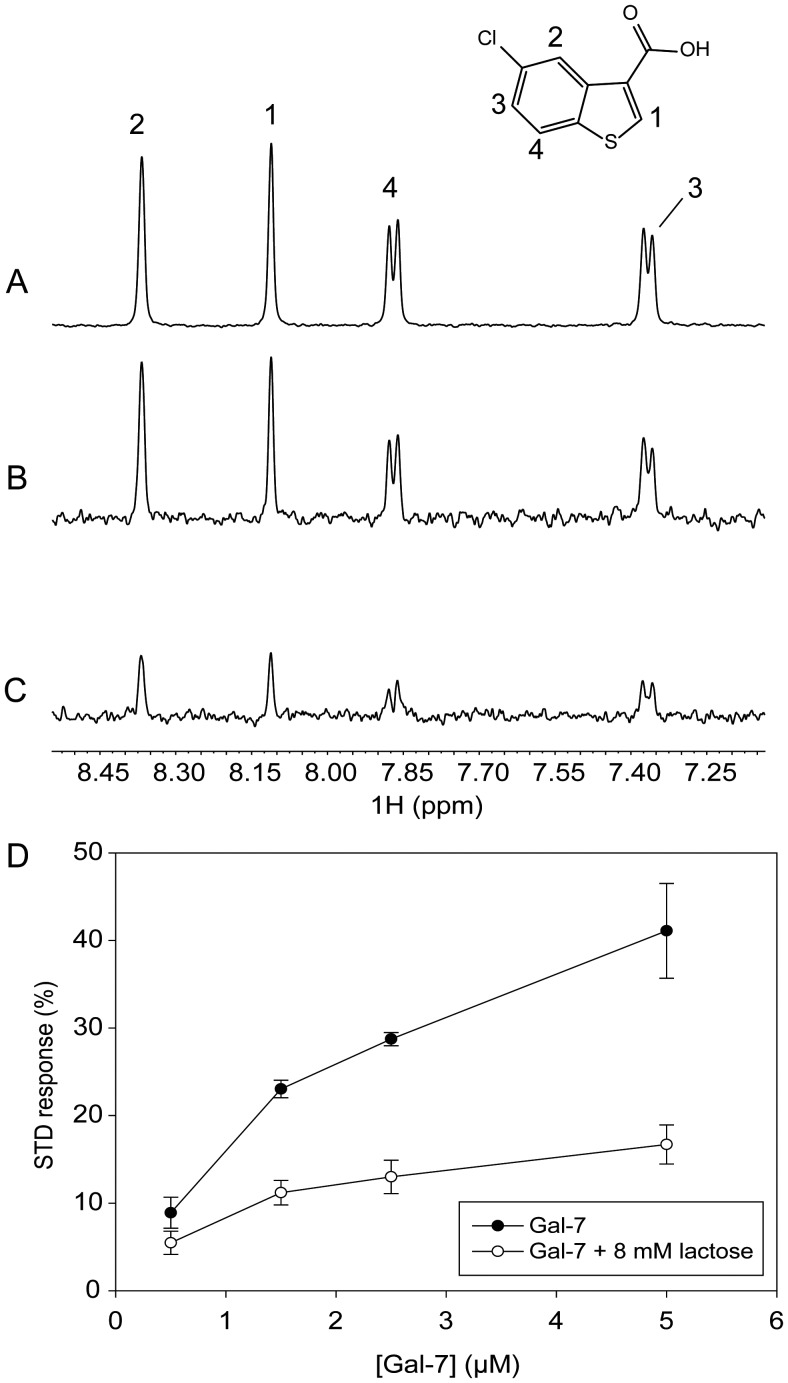
Lastly, we used STD NMR, a technique that is based on the transfer of magnetization from an irradiated protein to a weakly bound small molecule, i.e., with equilibrium dissociation constants in the micromolar to millimolar range (Meyer and Peters 2003; Roldós et al. 2011). For these STD NMR experiments, we used the small molecule probe 5-chloro-benzo[b]thio-phene-3-carboxylic acid (CBTC) that binds to the Gal-7 monomer at the dimer interface as a “spy” molecule to monitor its allosteric displacement by lactose binding. Binding of CBTC to Gal-7 is reflected in the increase of positive resonance intensity in the STD spectrum (Figure 8B). To demonstrate that CBTC binds at the Gal-7 dimer interface, we first acquired HSQC spectra of the monomeric 15N-Gal-3 carbohydrate-recognition domain, in the presence and absence of 500 μM CBTC. Supplementary data, Figure S1A and B, show that CBTC induces chemical shift changes of selective cross-peaks that correspond to residues located within a hydrophobic patch around K103 in a region that is structurally homologous to the Gal-7 dimer interface. Fittingly, at Gal-7 concentrations >100 μM where dimer is dominant, HSQC spectra of 15N-Gal-7 acquired in the presence and absence of 500 μM CBTC show no differences (data not shown), indicating that CBTC does not interact with Gal-7 dimers. We conclude, therefore, that CBTC binding is proportional to the amount of galectin monomer in solution. This concept is illustrated in Supplementary data Figure S2, which shows stronger binding of CBTC to the monomer state and weaker binding of CBTC to the dimer state.
Fig. 8.
1H NMR spectra of samples containing 500 µM CBTC in the presence of 1.5 µM Gal-7. (A) Reference spectrum (scaled 25%), with labels indicating the assignment of proton resonances. (B) STD spectrum. (C) STD spectrum in the presence of 8 mM lactose. (D) Plots showing STD responses for 500 µM CBTC vs. the concentration of Gal-7, before (filled circles) and after (open circles) addition of lactose to reach the concentration of 8 mM.
STD NMR data with CBTC (500 μM) in the presence of Gal-7 at a concentration of 1.5 μM, where monomers and dimers should be present in comparable amounts, are exemplified in Figure 8. This figure shows the chemical structure and reference NMR spectrum of CBTC (A, scaled 25%) with labels indicating resonance assignments, and STD NMR spectra of CBTC in the absence (B) and presence (C) of lactose. Figure 8D plots the percent STD effect with CBTC vs. the concentration of Gal-7 (500 nM–5 µM) in the absence (filled circles) and presence (open circles) of lactose. Because the actual amount of monomer in fact increases as the total concentration of Gal-7 increases, the STD effect also increases with the Gal-7 concentration. It is interesting to note that the mid-point of change on the lactose-free curve falls between 1 and 2 μM Gal-7, consistent with our FPLC-estimated value for the Gal-7 dimer Kd of 1.7 × 10−6 M. Moreover, because STD signals are significantly reduced in the presence of lactose at any concentration of Gal-7, we again conclude that binding of lactose stabilizes the dimer state, further supporting our previous results.
Further insight into ligand binding-induced long-range effects in Gal-7
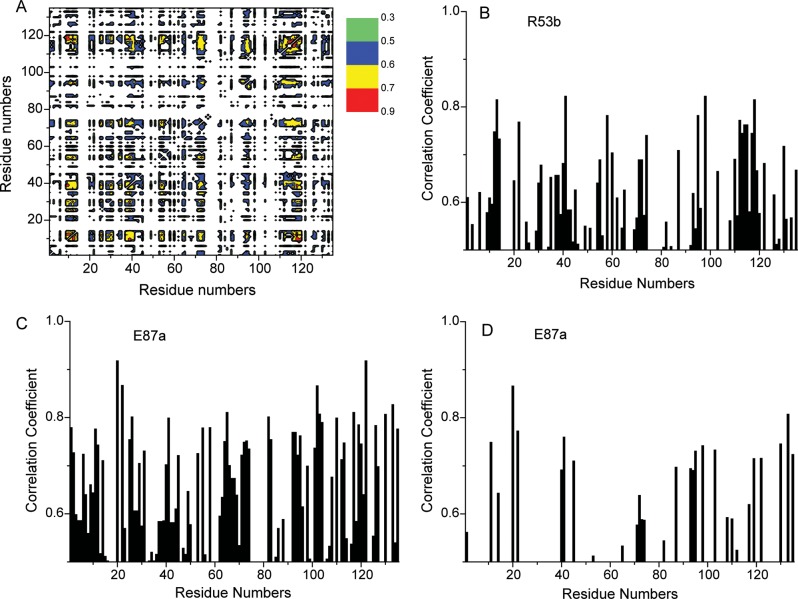
Because our HSQC data indicate cooperativity in carbohydrate ligand binding to Gal-7, there must be communication between ligand binding sites that should be transmitted from one residue to another and therefore should be related to motional coupling between/among residues. For insight into this idea, we turned again to MD simulations to assess motional coupling between residues by using covariance analysis of coordinate displacements averaged over simulation trajectories. Covariance maps (Figure 9) are expressed as mean-squared correlations from −1.0 to 1.0 and essentially describe linear correlations between pairs of Cα atoms as they fluctuate around their average positions during simulations. Covariance maps for the average of subunits of completely ligand-free Gal-7, the ligand-free subunit a in the half-loaded state and ligand-loaded subunit b in the half-loaded state are shown in Figure 9A–C, respectively. Although correlations can be positive or negative (corresponding to relative motions in the same or opposite directions, respectively) as denoted by colored contours in Figure 9, we focus discussion on the absolute value of the correlation. In the following analyses, the letters a and b refer to the two subunits of the half-loaded state of the dimer (PDB 4GAL), using the nomenclature adopted in Figure 9, that is, a for the ligand-free subunit and b for the lactose-bound subunit.
Fig. 9.
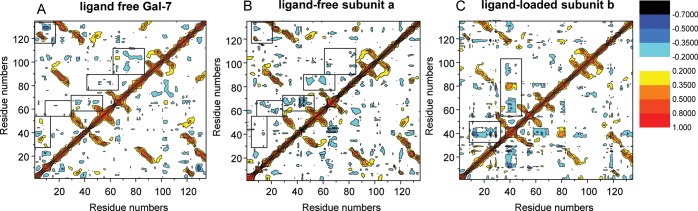
DCCM are shown as covariance maps for mean-squared motional correlations from −1.0 to 1.0. The plot essentially describes motional correlations between pairs of Cα atoms as they fluctuate around their average position during the MD simulation. Correlations can be positive (colored from yellow to red) or negative (colored from light blue to black) as relative motions move in the same or opposite directions, respectively. Covariance maps for the average of subunits of fully ligand-free Gal-7 (A), ligand-free subunit a in the half-loaded state (B) and ligand-loaded subunit b in the half-loaded state (C) are shown.
As expected, the most significant correlated motions in all the three instances are observed between residues from proximal β-strands within each β-sheet and between residues from sheets across the β-sandwich core (contours running orthogonal to the diagonal). Other significant motional correlations, however, are also apparent. The most interesting of these are within the lactose-binding site and at the dimer interface, as boxed-in in each covariance map. Aside from the expected correlations of residues between strands, we note that there are also strong correlations between β3-β4 and β5-β6 loops and residues within the carbohydrate-binding site, as well as between these loops and residues at the dimer interface. Ligand binding apparently promotes reorientation of residues and alteration of correlation patterns in both subunits (ligand-free and ligand-occupied) (Figure 9). These findings are supported by residue–residue (pairwise) interaction energies as presented in Supplemental data (Supplementary data, Figures S3 and S4), where we show that inter-residue van der Waals interactions (e.g., 15–20 and 88–98, and 90–95 and 104–113) are not as significant as electrostatic interactions, in particular between/among dimer interface residues (R14a-D94b, R20a-E87b, R20a-R22b, R22a-D103b, E87a-R22b, D94a-R14b, D95a-R14b, K98a-F135b and D103a-R133b). We believe that highly concerted motions within these regions may contribute to the energy transduction mechanism.
We also calculated residue–residue interaction energies using the method of Kong and Karplus (2007, 2009). This approach is based on calculating energy correlations between all interacting residue pairs in the MD ensemble of protein conformers where relative subunit motions do not influence intra-molecular pairwise interaction energies and are independent of the origin of the correlation. We focused our analysis on side-chain electrostatic interactions (Supplementary data, Figures S3 and S4) because they are long-range in nature, anisotropic, directionally more specific and can reflect small changes in the relative arrangements of residues vis-à-vis van der Waals and solvation energies. Figure 10A illustrates a projection of the interaction energy correlation matrix in residue space (residue correlation matrix), as suggested by Kong and Karplus (2007, 2009), for subunit b (ligand-bound subunit) for the half-loaded state of Gal-7 (for details, see Materials and methods). The residue correlation matrix shows the coupling between any two residues in the system. Although the average correlation coefficient per residue is decreased slightly upon ligand binding, individual coefficients increase in several regions, particularly for residues noted above in the proposed gradient. Long-range coupling must result from the extensive network of hydrogen bonds in β-sheets, long-range electrostatic effects and the overall β-sandwich structure of the protein.
Fig. 10.
Projections of the interaction energy correlation matrix on residue space are shown for ligand-loaded subunit b of the half-loaded state of Gal-7 (A), for residue R53 of subunit b of the half-loaded state (B) and for residue E87 of ligand-free subunit a within the same subunit (C) and with the opposing subunit b (D). Even with the various restrictions outlined in Materials and Methods, several thousand pairwise correlations, with correlation coefficients >0.5, were found for both ligand-free and ligand-loaded lectin.
We choose two residues to illustrate these long-range correlations, R53 (subunit b, Figure 10B) and E87 (subunit a, Figure 10C). R53 is at the ligand-binding site and interacts directly with lactose via a hydrogen bond. E87 is at the dimer interface and interacts with R20 via a salt bridge and with other surrounding interfacial residues (see Figure 6). The R53 projection (Figure 10B) indicates significant motional coupling with, e.g., residues G12-I13-R14, R22, E41, D55, E58, V60, R74, D95, K98, D103, as well as with much of the L9 loop (residues 110–119) and D130, R133. Binding of lactose to subunit b impacts on correlated motions not only within the ligand-binding site but also through the domain to the back side of the β-sandwich and residues involved in inter-subunit interactions.
For E87 (unbound subunit a), the projection is shown in two parts. Figure 10C shows correlated motions within the same subunit a, while Figure 10D shows correlated motions with residues of lactose-bound subunit b. From Figure 10C, it is apparent that there are numerous correlated motions between E87 and other residues within the same subunit. Of particular note are correlations with residues at the dimer interface (12–14, 93–98, 103, 110–119 and 133). Correlated motions with residues from opposing subunit b are more limited; nonetheless, they are significant (Figure 10D) and include interfacial residues G12, R14, R20, R22 and residues 92–100 (loop L7-β8) and 130–135 (β11), as well as residues E40, E41, S44 and E72 from the lactose-binding site and additional residues F86 and 117–122 (β10). Changes in dynamics at the dimer interface likely contribute to ligand binding-induced increase in dimer stability. Correlated motions of dimer interface residues that are significantly influenced by ligand binding are nicely exemplified by salt-bridged residues R20a and E87b (or R20b and E87a—see Figure 6), which upon lactose binding reorient significantly to a more thermodynamically favorable state (see Supplemental Results, Supplementary data, Figure S5). In general, binding of lactose to Gal-7 does not greatly alter the network of correlated motions between/among residues, but rather modulates or attenuates these correlations via effects on side-chain (and backbone) flexibility, which is increased overall upon ligand binding.
Discussion
Evolutionary divergence has led to the establishment of three types of structural organization of galectins and further phylogenetic diversification in two subgroups. The use of ligand derivatives (chemical mapping), profiling of binding specificities by glycans/cell reactivity and molecular modeling based on X-ray structures has already delineated differences for proto-type galectins (Solís et al. 1996; Leonidas et al. 1998; Hirabayashi et al. 2002; López-Lucendo et al. 2004; Wu et al. 2007; Habermann et al. 2011). The recent introduction of isotopically labeled galectins to structural analysis in solution has made it possible to study dynamic aspects of the protein (Diehl et al. 2009, 2010; Nesmelova et al. 2010). This question, together with the search for long-range effects upon ligand binding, has been addressed herein.
In the present study, we report that the binding of lactose to Gal-7 indeed has a long-range impact on protein thermodynamics and conformational dynamics up to the dimer interface and, further away, between binding sites. Using a two-site model for curve fitting, we provide evidence for lactose binding with positive cooperativity, such that binding of the first ligand (K1 = 0.9 ± 0.6 × 103 M−1) enhances binding of the second (K2 = 3.4 ± 0.8 × 103 M−1). In structural terms, analysis of MD simulations on the half-loaded state indicates that significant changes occur in the ligand-free subunit a when lactose is bound to subunit b. Moreover, internal motions in Gal-7 are on average increased overall upon ligand binding, reflecting increased conformational entropy. In global terms, similar observations have been made for Gal-1 (Nesmelova et al. 2010) and Gal-3 (Diehl et al. 2009, 2010), where lactose binding to both galectins was reported to result in generally increased backbone internal motions. Thermodynamically, the TΔS term appears higher for Gal-7 when binding N-acetyllactosamine, compared with Gal-1 and -3 (Ahmad et al. 2002; Dam et al. 2005). Of particular note, initial data on Gal-3 and also Gal-1 indicate that this effect appears to depend on the nature of the ligand (Diehl et al. 2010; Miller et al. 2011). Thus, variation of the binding signature or ligand contact pattern at the lectin site appears capable of modulating conformational dynamics, warranting systematic assessments.
In detail, we observed that the binding of lactose to Gal-7 alters the lectin conformation and dynamics within the ligand-binding site, as well as through an apparent internal gradient from the ligand-binding site to the dimer interface and beyond. The greatest effects are observed in the 50–58 and 62–70 loops (whose residues interact directly with the ligand), the 5-stranded β-sheet at the backside of the lactose-binding site (including the region involved in dimerization of Gal-7) and loops (especially residues 9–14 and 110–116) down to the dimer interface. The most affected residues are highlighted on the structure of Gal-7 in Figure 11 (see also Supplementary data). The 9–14 loop (part of the Gal-7 dimer interface) connects β-strand 1 on the lactose-binding face with β-strand 2 on the backside of Gal-1, and the 110–116 loop runs from the backside of the lactose-binding site, down one side of the β-sandwich to β-strand 10, again connecting β-strands on both sides of the β-sandwich.
Fig. 11.
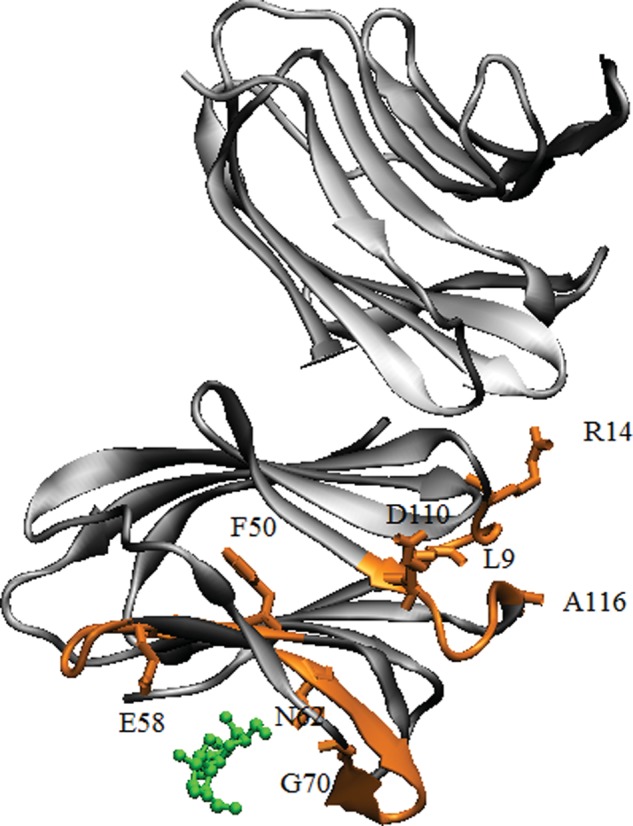
The binding of lactose to Gal-7 induces alterations in the protein conformation and dynamics within the ligand binding site, as well as through an internal gradient from the ligand-binding site to the dimer interface, as highlighted in orange. Lactose is shown in green.
Some of the most significant changes in Gal-1 are in fact also noted at residues 110–116, a loop that contains residues R110 and R112, which in Gal-7 are affected by ligand loading. While these and other conformational and dynamical changes with Gal-7 subunits are similar to those observed with Gal-1 subunits (Nesmelova et al. 2010), ligand binding to the Gal-1 dimer was found to occur with negative cooperativity, with binding of the first ligand in fact reducing affinity for the second ligand. This stands in stark contrast to positive cooperativity with Gal-7 as observed here, and suggests that the quarternary structures of Gal-1 and -7 dimers themselves, which are quite different in these two prototype galectins (Nesmelova et al. 2008a), may actually direct how the ligand binding-induced signal is channeled into cooperativity in ligand binding.
Since our modeling work suggested an increase in inter-residue interaction energies at the dimer interface, we performed several experimental studies (FPLC gel filtration, hetero-FRET, CD-based thermal denaturation and STD NMR) with Gal-7 in the absence and presence of lactose. In all cases, the presence of a saturating amount of lactose resulted in stabilization of the Gal-7 dimer state. Although the presence of lactose significantly increased the dimer population, we could only estimate that the Kd for the lactose-loaded Gal-7 was <<0.4 × 10−6 M, the maximum possible Kd, conservatively estimated taking into account the law of mass action, being 0.013 × 10−6 M. This would suggest that ΔΔG is changed by a minimum of –2.9 kcal/mol. Moreover, even though our MD-calculated interaction energies cannot be compared directly with these experimentally determined free energies due to the absence of contributions from entropy terms and over-estimation of energies for various reasons, e.g., scaling factors (Nesmelova et al. 2008b), the trends are the same, i.e., ΔΔG values are negative, and therefore dimer stability is enhanced. Of note, this behavior is different for Gal-1, which showed no difference in the elution profile under identical conditions. Although difficult to quantify, it is apparent that Kd values for Gal-1 dimer dissociation in the presence or absence of lactose must be considerably less than those for Gal-7, i.e., Gal-1 dimers are more thermodynamically stable than Gal-7 dimers.
In aggregate, our study with Gal-7 detected significant ligand binding-induced long-range effects, particularly in terms of changes to conformational dynamics. In view of the available evidence from Gal-1 and -3 (Umemoto et al. 2003; Diehl et al. 2009, 2010; Nesmelova et al. 2010) and phylogenetic considerations within this protein family (Houzelstein et al. 2004; Kaltner and Gabius 2012), the present data provide further insight into ligand binding-induced differential flexibility among galectins, a starting point for structure-activity correlations by mutational engineering. Conversely, the analysis of galectin-derived peptides confined to ligand-contacting sequence stretches (Moise et al. 2011) will help to dissect contributions of effects in immediate vicinity and more distant regions. In addition, such comparisons will be possible to lectins in terms of folding, where Ca2+ is involved in ligand contact/structural stabilization (Gabius 2011), and to discern the principles on how a common folding in different lectins (i.e., the β-sandwich) responds to ligand binding. Having delineated the impact of lactose binding, it will also be intriguing to study the effects of cognate peptide motifs from protein counter-receptors.
Materials and methods
Production of labeled proteins
Production of 15N-labeled human Gal-7 (15N-Gal-7) and controls for purity and activity have been described elsewhere (Nesmelova et al. 2012). Production of 15N-labeled truncated form of human galectin-3 with collagenase followed the protocol described previously (Kübler et al. 2008).
For the FRET labeling procedure, freeze-dried Gal-7 was dissolved in 0.02 M potassium phosphate buffer, pH = 7.0, with 0.01 M lactose and 2 mM dithiothreitol (DTT). After 1 h incubation, the solution was desalted to remove excess of DTT, and divided into two moieties for labeling with donor (EDANS-C2-maleimide) and acceptor (DABCYL-C2-maleimide). To increase solubility of DABCYL-C2-maleimide, 5% vol/vol of dimethylformamide (DMF) was added to the stock solution. Upon labeling, the ratio protein:label was 1:4, and samples were incubated at 4°C overnight. Excess of free label was removed thereafter by changing buffer using Amicon spin-concentrators (MWCO 3000). Additionally, to remove possibly degraded and aggregated protein, prepared samples were centrifuged at 90,000 × g for 30 min. The extent of labeling (the fraction of labeled Gal-7) was determined via comparison of protein concentration and probe concentration in the sample of labeled protein. Gal-7 concentration was measured with Bradford assay (BioRad Hercules, CA, USA). The concentration of probes in the sample of labeled protein was measured in denaturing buffer containing 0.05 M MOPS, 10% v/v glycerol, 5% v/v ethanol and 2% w/w sodium dodecyl sulfate. Under these conditions, the extinction coefficient for EDANS and DABCYL was found as ϵ336 = 6280 M−1 cm−1 and ϵ450 = 20,800 M−1 cm−1, accordingly. Initial concentrations of the EDANS and DABCYL probes were determined by absorbance of DABCYL in methanol at 428 nm (ϵ = 22,330 M−1 cm−1, AnaSpec) and EDANS in water (ϵ = 6100 M−1 cm−1, Hudson and Weber 1973). The extent of labeling was 17% for DABCYL-labeled Gal-7 and 65–85% for EDANS-labeled Gal-7. To remove the excess of lactose, the buffer was changed with spin concentrators (Amicon) and desalting columns (Pierce). In FRET experiments the concentration of donor-labeled Gal-7 was maintained at 1 μM.
NMR spectroscopy
Uniformly 15N-labeled Gal-7 was dissolved at a concentration of 1 mg/mL (67 μM monomer) in 20 mM potassium phosphate buffer at pH 7.0, made up using a 95% H2O/5% D2O mixture containing 2 mM DTT. 1H–15N HSQC NMR experiments were used to investigate binding of lactose. 1H and 15N resonance assignments for recombinant Gal-7 had previously been reported (Nesmelova et al. 2012).
NMR experiments were carried out at 30°C on a Varian Unity Inova 600-MHz spectrometer equipped with an H/C/N triple-resonance probe and x/y/z triple-axis pulse field gradient unit. A gradient sensitivity-enhanced version of two-dimensional 1H–15N HSQC was applied with 256 (t1) × 2048 (t2) complex data points in nitrogen and proton dimensions, respectively. Raw data were converted and processed by using NMRPipe (Delaglio et al. 1995) and were analyzed by using NMRview (Johnson and Blevins 1994).
In 1H–15N HSQC experiments on proteolytically truncated Gal-3, samples contained 0.2 mM uniformly 15N-labeled protein in 20 mM phosphate buffer, pH 7.4 in H2O, plus 10% D2O for field-frequency lock. Spectra were recorded at 298 K in a Bruker Avance 600-MHz spectrometer equipped with a cryogenically cooled HCN probe with z-axis gradients and processed with MestReNova. Resonance assignments are available at BMRB, entry 15705.
Determination of the binding constants by NMR
Monte Carlo calculations were used to determine values of K1 and K2 from titration curves acquired using HSQC spectra on 15N-labeled Gal-7 as a function of lactose concentration. Calculations using one-site and two-site ligand-binding models were performed as previously described (Nesmelova et al. 2010; Miller et al. 2011) with 20 individual 1H and 15N resonances that were most chemically shifted during the titration.
STD-NMR studies
Samples for STD experiments were prepared with 500 µM CBTC as a molecular probe (Maybridge, Cambridge, UK) and Gal-7 concentrations around its apparent dimer KD value (0.5, 1.5, 2.5 and 5.0 μM) in fully deuterated 20 mM phosphate buffer at p2H 7.4. The effect of lactose was analyzed by repeating the experiments after the addition of 8 mM lactose to the samples.
STD-NMR data were collected at 298 K on a Bruker Avance DRX 500 MHz spectrometer equipped with a 5 mm inverse probe head. The protein was saturated on-resonance at 0.8 ppm and off-resonance at 100 ppm with a train of Gaussian-shaped pulses of 50 ms each and a total saturation time of 2 s. Experiments were performed by triplicate, rendering reproducible results.
Gel filtration FPLC studies
Gel filtration was performed with a Superose 12 10/300 GL column (void volume: 7.2 mL; Amersham Biosciences AB, Uppsala, Sweden) equilibrated at 30°C with 20 mM potassium phosphate buffer, pH 7.0, containing 2 mM DTT and 0.02% NaN3, in the absence or presence of 10 mM lactose. The flow rate was 0.5 mL/min, and the elution was monitored at 280 and 215 nm simultaneously using an ÄKTA Purifier equipment (GE Healthcare Lifesciences, Diegen, Belgium). In each experiment, the concentration of the eluted protein was calculated using the dilution factor estimated from the ratio of width at half-height of the peak to the injection volume (100 μL). Blue dextran (2000 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa) and cytochrome c (12.4 kDa) were fractionated for calibration under similar conditions.
Circular dichroism
CD spectra were acquired at 20 and 30°C in a J-810 spectropolarimeter equipped with a Peltier temperature control system, using a band width of 1 nm and a response time of 2 s. Far UV spectra were recorded in 0.2/0.1 cm path length quartz cells at a protein concentration of 0.1–1.25 mg/mL (6.6–83 μM monomer), while near-UV spectra were registered at 0.3–4.2 mg/mL (20–280 μM monomer) in 1/0.2 cm pathlength cells. Spectra were recorded in 20 mM potassium phosphate buffer, pH 7.0, containing 2 mM DTT, in the absence or presence of 10 mM lactose. For all the spectra, the corresponding buffer baseline was subtracted. Thermal denaturation experiments were carried out by increasing the temperature from 20 to 80°C at a scanning rate of 0.33°C/min. Variations in ellipticity at a given wavelength were monitored every 0.2°C. Thermal denaturation profiles were described in terms of the following sigmoidal function:
where T is the absolute temperature, T1/2 is the half transition temperature, R is the gas constant, A is the temperature constant accounting for the variation of the ratio between the native and denatured states and ΘD (T) and ΘN (T) are the ellipticity of the denatured and native states at temperature T. ΘD and ΘN were approximated as linear functions of temperature, as described previously (Campanero-Rhodes et al. 2006).
Hetero-FRET measurements
Fluorescence of EDANS-Gal-7 was excited with the third harmonics of a passively Q-switched microchip YAG laser (SNV-20F-100, Teem Photonics Meylan, France), operated at a pulse repetition rate of 20 kHz and selected with a 409 nm long-pass glass filter. To avoid anisotropy effects, fluorescence was passed through a polarizer oriented at the magic angle. Fluorescence signals were detected after every laser shot with a photomultiplier module Hamamatsu H5773-20 and acquired with a transient digitizer (Acqiris DC252, Agilent Palo Alto, CA, USA) with a sampling resolution of 0.125 ns. The instrument response function (IRF) was acquired with scattered light at the same instrument settings as in the fluorescence measurement, except that there was no emission filter and the emission polarization was vertical. The sample was placed in thermostated cuvette, and all experiments were performed at 30°C. Analysis of the data was performed as described elsewhere (Agafonov et al. 2009). In short, traces of donor fluorescence were fitted as one-exponential decay to examine the effect of dimer formation. The decrease in apparent lifetime of donor fluorescence with an increase of acceptor concentration unambiguously confirms a FRET effect between donor and acceptor in formed dimers. The mole fraction of dimers with donor–acceptor pair was determined in two steps. First, fluorescent traces of donor-only-labeled Gal-7 and donor–acceptor-labeled Gal-7 (sub-stoichiometric acceptor concentrations) were fitted as two-exponential decay (Agafonov et al. 2009) to characterize donor fluorescence. Second, fluorescent traces of donor in the presence of acceptor ([acceptor] > [donor]) were fitted considering FRET between donor and acceptor, using parameters of donor fluorescence as determined in the first step. The Foerster radius R0 = 4 nm for EDANS–DABCYL pair (Agafonov et al. 2009). All observed fluorescent traces were fitted globally, including samples with different lactose concentrations. In these fits, amplitudes and lifetimes of donor fluorescence were linked and not varied. The interprobe distance and the Gaussian width of the interprobe distance distribution were linked and varied. The fraction of donor-only-labeled Gal-7 was not linked and varied. These fits revealed an interprobe distance of 4.2 nm, in excellent agreement with the Cα—Cα distance in the X-ray structure (3.8 nm, Leonidas et al. 1998).
MD simulations
The coordinates of ligand-free Gal-7 and complexes of Gal-7 with one lactose molecule bound and with two galactoses bound were obtained from the PDB (codes are 1BKZ, 4GAL and 2GAL, respectively) (Bernstein et al. 1977). In all instances, Gal-7 is a dimer. In the crystallographic structure of the complex with one lactose molecule, only subunit b hosts a molecule of lactose, while subunit a is free of ligand. In addition, one trajectory was created where lactose was removed from the complex, i.e., from subunit b.
MD simulations were performed by using the NAMD 2.6 (Phillips et al. 2005) software with CHARMM force field parameters for the protein, CARBOHYDRATE parameters for lactose and galactose and TIP3P for water molecules. Simulations were carried out under sothermal-isobaric (NPT) ensemble conditions with periodic boundary conditions and Particle Mesh Ewald (PME) electrostatics using a 2 fs time step. The Langevin piston method was used to maintain a constant pressure of 1 atm with a piston period of 100 fs and a piston decay of 50 fs. The temperature was maintained at 300 K. The dimer of Gal-7 without ligand or in complexes with ligand was solvated in a box of 61 × 71 × 81 Å3. Following initial minimization, heating and equilibration stages, the production phase of each MD simulation ran for 20 ns. Coordinates were saved every 10 ps for further analysis. The overall stability of each simulation system was evaluated by monitoring the time evolution of the RMSD of each Cα atom with respect to the initial structure. Trajectories were analyzed using VMD (Humphrey et al. 1996) and CHARMM (MacKerell et al. 1998) program packages. All molecular images were prepared with VMD. Because relative movements of subunits in the dimer can obscure analysis of intra-subunit correlated motions, rigid-body rotational and translational motions were accounted for by fitting frames on the reference structure and by performing superposition and RMSF calculations individually for each subunit.
Binding energy calculations
Binding energies were calculated as the difference between the energy of the complex and the energy of the individual components using the MM/PBSA approach (Kollman et al. 2000).
where EvdW stands for the van der Waals interaction energy; EPB stands for the electrostatic interaction energy and polar contribution of solvation energy and ESASA characterizes the nonpolar contribution to the solvation effect.
Dynamic cross-correlation matrix
Motional correlations between/among residues were analyzed by examining the dynamic cross-correlation map (DCCM) of Cα atoms for lactose-loaded and ligand-free Gal-7. Protein structures from the trajectory were superimposed to a reference structure in order to remove overall translational and rotational motions of the protein.
The cross-correlation matrix element Cij of atoms i and j was calculated as:
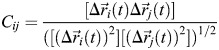 |
where < > denotes an MD-averaged quantity. The displacement from the average MD position of atom i is given by: 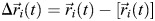 .
.
Interaction energy correlation
Interaction energies between residue pairs were calculated as a function of time in the 20 ns MD simulation. Only residue–residue interactions with average interaction energies >1 kcal/mol were analyzed (Kong and Karplus 2007). The correlation between two sets of residue–residue interaction energies i, j and k, l is defined as:
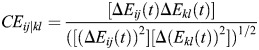 |
where
and 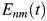 is the electrostatic energy between protein residues m and n.
is the electrostatic energy between protein residues m and n.
We also used the method of Kong and Karplus (2007, 2009), which is based on calculating energy correlations between all interacting residue pairs in the MD ensemble of protein conformers. The resulting interaction correlation matrix complements the covariance map approach, with the added advantage that relative subunit motions do not influence intra-molecular pairwise interaction energies. To create this interaction correlation matrix, we first calculated electrostatic interaction energies between all residue pairs as a function of time during the MD trajectory and then determined equal-time correlations for the interaction energies. Thermal noise was reduced by averaging over a time window of 100 ps, and we excluded sequential residue pairs and those pairs with average interaction energies less than 1 kcal/mol.
To visualize these pairwise correlations, we modified a projection of the interaction energy correlation matrix in residue space (residue correlation matrix) as suggested by Kong and Karplus (2007, 2009), and created a matrix in which each element was equal to the average absolute value of the correlation coefficient for any pair of residues. For example, if the interaction energy correlation coefficient for residue pairs 20.87 and 49.51 (i.e., 20.87|49.51) is 0.7 and that for pairs (103.87|51.64) is 0.5, then the projection on the matrix for residues 87 and 51 would be 0.6. Because each column and row in the correlation matrix represents a specific residue in the protein, we end up with a normalized 135 × 135 matrix for the monomer and a 270 × 270 matrix for the dimer. This matrix then represents pairwise correlations between distant residues, independently of the origin of the correlation.
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by a research grant from the National Institutes of Health (CA 096090 to K.H.M.), the RAS program “Molecular and Cellular Biology” RFBR grant (No. 12 04 31360 to E.E.), the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no 260600 (“GlycoHIT”), grants CTQ2009-08536 and BFU2009-10052 and a FPI PhD fellowship to M.A.B. from the Spanish Ministry of Science and Innovation and the CIBER of Respiratory Diseases (CIBERES), an initiative from the Spanish Institute of Health Carlos III (ISCIII). E.E. was supported by a Travel Grant from the Minnesota Supercomputing Institute (University of Minnesota) during her stay in the research lab of Prof. K.H. Mayo. I.N. was supported in the Mayo lab by National Institutes of Health Hematology Training Grant (HL 07062).
Conflict of interest
None declared.
Abbreviations
15N-Gal-7, 15N-enriched Gal-7; 15N-Gal-7, 15N-labeled human Gal-7; CBTC, 5-chloro-benzo[b]thio-phene-3-carboxylic acid; CD, circular dichroism; DCCM, dynamic cross-correlation map; FPLC, fast protein liquid chromatography; Gal-1, galectin-1; Gal-7, galectin-7; IRF, instrument response function; MM/PBSA, molecular mechanics/Poisson–Boltzmann surface area; NMR, nuclear magnetic resonance; PDB, protein data bank; RMSF, root mean-square fluctuation; STD, saturation transfer difference.
Supplementary Material
Acknowledgements
The authors thank the Minnesota Supercomputing Institute (University of Minnesota) for providing computer resources and Drs B. Friday and W. Notelecs for helpful discussions.
References
- Agafonov RV, Negrashov IV, Tkachev YV, Blakely SE, Titus MA, Thomas DD, Nesmelov YE. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. Proc Natl Acad Sci USA. 2009;106:21625–21630. doi: 10.1073/pnas.0909757106. doi:10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N, Gabius H-J, Kaltner H, André S, Kuwabara I, Liu F-T, Oscarson S, Norberg T, Brewer CF. Thermodynamic binding studies of cell surface carbohydrate epitopes to galectins-1, -3, and -7: Evidence for differential binding specificities. Can J Chem. 2002;80:1096–1104. doi:10.1139/v02-162. [Google Scholar]
- André S, Kaltner H, Furuike T, Nishimura S, Gabius H-J. Persubstituted cyclodextrin-based glycoclusters as inhibitors of protein-carbohydrate recognition using purified plant and mammalian lectins and wild-type and lectin-gene-transfected tumor cells as targets. Bioconjug Chem. 2004;15:87–98. doi: 10.1021/bc0340666. doi:10.1021/bc0340666. [DOI] [PubMed] [Google Scholar]
- André S, Pieters RJ, Vrasidas I, Kaltner H, Kuwabara I, Liu F-T, Liskamp RM, Gabius H-J. Wedgelike glycodendrimers as inhibitors of binding of mammalian galectins to glycoproteins, lactose maxiclusters, and cell surface glycoconjugates. Chem Bio Chem. 2001;2:822–830. doi: 10.1002/1439-7633(20011105)2:11<822::AID-CBIC822>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Bernerd F, Sarasin A, Magnaldo T. Galectin-7 overexpression is associated with the apoptotic process in UVB-induced sunburn keratinocytes. Proc Natl Acad Sci USA. 1999;96:11329–11334. doi: 10.1073/pnas.96.20.11329. doi:10.1073/pnas.96.20.11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rogers JR, Kennard O, Shimanouchi T, Tatsumi M. The protein data bank: A computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. doi:10.1016/S0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Campanero-Rhodes MA, Menéndez M, Sáiz JL, Sanz L, Calvete JJ, Solís D. Zinc ions induce the unfolding and self-association of boar spermadhesin PSP-I, a protein with a single CUB domain architecture, and promote its binding to heparin. Biochemistry. 2006;45:8227–8235. doi: 10.1021/bi052621g. doi:10.1021/bi052621g. [DOI] [PubMed] [Google Scholar]
- Cooper DNW. Galectinomics: Finding themes in complexity. Biochim Biophys Acta. 2002;1572:209–231. doi: 10.1016/s0304-4165(02)00310-0. doi:10.1016/S0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- Dam TK, Gabius H-J, André S, Kaltner H, Lensch M, Brewer CF. Galectins bind to the multivalent glycoprotein asialofetuin with enhanced affinities and a gradient of decreasing binding constants. Biochemistry. 2005;44:12564–12571. doi: 10.1021/bi051144z. doi:10.1021/bi051144z. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Diehl C, Engström O, Delaine T, Håkansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ, Akke M. Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3. J Am Chem Soc. 2010;132:14577–14589. doi: 10.1021/ja105852y. doi:10.1021/ja105852y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl C, Genheden S, Modig K, Ryde U, Akke M. Conformational entropy changes upon lactose binding to the carbohydrate recognition domain of galectin-3. J Biomol NMR. 2009;45:157–169. doi: 10.1007/s10858-009-9356-5. doi:10.1007/s10858-009-9356-5. [DOI] [PubMed] [Google Scholar]
- Gabius H-J. The how and why of Ca2+ involvement in lectin activity. Trends Glycosci Glycotechnol. 2011;23:168–177. doi:10.4052/tigg.23.168. [Google Scholar]
- Gabius H-J, André S, Jiménez-Barbero J, Romero A, Solís D. From lectin structure to functional glycomics: Principles of the sugar code. Trends Biochem Sci. 2011;36:298–313. doi: 10.1016/j.tibs.2011.01.005. doi:10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Habermann FA, André S, Kaltner H, Kübler D, Sinowatz F, Gabius H-J. Galectins as tools for glycan mapping in histology: Comparison of their binding profiles to the bovine zona pellucida by confocal laser scanning microscopy. Histochem Cell Biol. 2011;135:539–552. doi: 10.1007/s00418-011-0814-2. doi:10.1007/s00418-011-0814-2. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J, Hashidate T, Arata Y, Nishi N, Nakamura T, Hirashima M, Urashima T, Oka T, Futai M, Müller WE, et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim Biophys Acta. 2002;1572:232–254. doi: 10.1016/s0304-4165(02)00311-2. doi:10.1016/S0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Gonçalves IR, Fadden AJ, Sidhu SS, Cooper DNW, Drickamer K, Leffler H, Poirier F. Phylogenetic analysis of the vertebrate galectin family. Mol Biol Evol. 2004;21:1177–1187. doi: 10.1093/molbev/msh082. doi:10.1093/molbev/msh082. [DOI] [PubMed] [Google Scholar]
- Hudson EN, Weber G. Synthesis and characterization of two fluorescent sulfhydryl reagents. Biochemistry. 1973;12:4154–4161. doi: 10.1021/bi00745a019. doi:10.1021/bi00745a019. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J Mol Graph. 1996;141:33–38. doi: 10.1016/0263-7855(96)00018-5. doi:10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. doi:10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kaltner H, Gabius H-J. A toolbox of lectins for translating the sugar code: The galectin network in phylogenesis and tumors. Histol Histopathol. 2012;27:397–416. doi: 10.14670/HH-27.397. [DOI] [PubMed] [Google Scholar]
- Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. doi:10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Kong Y, Karplus M. The signaling pathway of rhodopsin. Structure. 2007;15:611–623. doi: 10.1016/j.str.2007.04.002. doi:10.1016/j.str.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Kong Y, Karplus M. Signaling pathways of PDZ2 domain: A molecular dynamics interaction correlation analysis. Proteins. 2009;74:145–154. doi: 10.1002/prot.22139. doi:10.1002/prot.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopitz J, André S, von Reitzenstein C, Versluis K, Kaltner H, Pieters RJ, Wasano K, Kuwabara I, Liu F-T, Cantz M, et al. Homodimeric galectin-7 (p53-induced gene 1) is a negative growth regulator for human neuroblastoma cells. Oncogene. 2003;22:6277–6288. doi: 10.1038/sj.onc.1206631. doi:10.1038/sj.onc.1206631. [DOI] [PubMed] [Google Scholar]
- Kübler D, Hung C-W, Dam TK, Kopitz J, André S, Kaltner H, Lohr M, Manning JC, He L, Wang H, et al. Phosphorylated human galectin-3: Facile large-scale preparation of active lectin and detection of structural changes by CD spectroscopy. Biochim Biophys Acta. 2008;1780:716–722. doi: 10.1016/j.bbagen.2008.01.018. doi:10.1016/j.bbagen.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Kuwabara I, Kuwabara Y, Yang RY, Schuler M, Green DR, Zuraw BL, Hsu DK, Liu F-T. Galectin-7 (PIG1) exhibits pro-apoptotic function through JNK activation and mitochondrial cytochrome c release. J Biol Chem. 2002;277:3487–3497. doi: 10.1074/jbc.M109360200. doi:10.1074/jbc.M109360200. [DOI] [PubMed] [Google Scholar]
- Leonidas DD, Vatzaki EH, Vorum H, Celis JE, Madsen P, Acharya KR. Structural basis for the recognition of carbohydrates by human galectin-7. Biochemistry. 1998;37:13930–13940. doi: 10.1021/bi981056x. doi:10.1021/bi981056x. [DOI] [PubMed] [Google Scholar]
- López-Lucendo MF, Solís D, André S, Hirabayashi J, Kasai K, Kaltner H, Gabius H-J, Romero A. Growth-regulatory human galectin-1: Crystallographic characterisation of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J Mol Biol. 2004;343:957–970. doi: 10.1016/j.jmb.2004.08.078. doi:10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics. Studies of proteins. J Phys Chem. 1998;B102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Madsen P, Rasmussen HH, Flint T, Gromov P, Kruse TA, Honoré B, Vorum H, Celis JE. Cloning, expression, and chromosome mapping of human galectin-7. J Biol Chem. 1995;270:5823–5829. doi: 10.1074/jbc.270.11.5823. doi:10.1074/jbc.270.11.5823. [DOI] [PubMed] [Google Scholar]
- Magnaldo T, Bernerd F, Darmon M. Galectin-7, a human 14-kDa S-lectin, specifically expressed in keratinocytes and sensitive to retinoic acid. Dev Biol. 1995;168:259–271. doi: 10.1006/dbio.1995.1078. doi:10.1006/dbio.1995.1078. [DOI] [PubMed] [Google Scholar]
- Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42:864–890. doi: 10.1002/anie.200390233. doi:10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- Miller MC, Ribeiro JP, Roldós V, Martín-Santamaría S, Cañada FJ, Nesmelova I, André S, Pang M, Klyosov A, Baum LG, et al. Structural aspects of binding of α-linked digalactosides to human galectin-1. Glycobiology. 2011;21:1627–1641. doi: 10.1093/glycob/cwr083. doi:10.1093/glycob/cwr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise A, Andre' S, Eggers F, Krzeminski M, Przybylski M, Gabius H-J. Toward bioinspired galectin mimetics: identification of ligand-contacting peptides by proteolytic-excision mass spectrometry. J Am Chem Soc. 2011;133:14844–14847. doi: 10.1021/ja201967v. doi:10.1021/ja201967v. [DOI] [PubMed] [Google Scholar]
- Morris S, Ahmad N, Andre S, Kaltner H, Gabius H-J, Brenowitz M, Brewer F. Quaternary solution structures of galectins-1, -3, and -7. Glycobiology. 2004;14:293–300. doi: 10.1093/glycob/cwh029. doi:10.1093/glycob/cwh029. [DOI] [PubMed] [Google Scholar]
- Nesmelova IV, Berbís MA, Miller MC, Cañada FJ, André S, Jiménez-Barbero J, Gabius H-J, Mayo KH. 1H, 13C, and 15N backbone and side-chain chemical shift assignments for the 31 kDa human galectin-7 (p53-induced gene 1) homodimer, a pro-apoptotic lectin. Biomol NMR Assign. 2012;6:127–129. doi: 10.1007/s12104-011-9339-9. [DOI] [PubMed] [Google Scholar]
- Nesmelova IV, Dings RPM, Mayo KH. “Understanding galectin structure-function relationships to design effective antagonists”. In: Klyosov A, Witczak ZJ, Platt D, editors. “Galectins”. New York: Oxford University Press; 2008a. pp. 33–69. [Google Scholar]
- Nesmelova IV, Ermakova E, Daragan VA, Pang M, Menéndez M, Lagartera L, Solís D, Baum LG, Mayo KH. Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity. J Mol Biol. 2010;397:1209–1230. doi: 10.1016/j.jmb.2010.02.033. doi:10.1016/j.jmb.2010.02.033. [DOI] [PubMed] [Google Scholar]
- Nesmelova IV, Sham Y, Gao J, Mayo KH. CXC-chemokines associate with CC-chemokines to form mixed heterodimers: RANTES and PF4 monnomers associate as CC-type heterodimers. J Biol Chem. 2008b;283:24155–24166. doi: 10.1074/jbc.M803308200. doi:10.1074/jbc.M803308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. doi:10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. doi:10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Remmelink M, de Leval L, Decaestecker C, Duray A, Crompot E, Sirtaine N, André S, Kaltner H, Leroy X, Gabius H-J, et al. Quantitative immunohistochemical fingerprinting of adhesion/growth-regulatory galectins in salivary gland tumours: Divergent profiles with diagnostic potential. Histopathology. 2011;58:543–556. doi: 10.1111/j.1365-2559.2011.03776.x. doi:10.1111/j.1365-2559.2011.03776.x. [DOI] [PubMed] [Google Scholar]
- Roldós V, Cañada FJ, Jiménez-Barbero J. Carbohydrate-protein interactions: A 3D view by NMR. Chem Biochem. 2011;12:990–1005. doi: 10.1002/cbic.201000705. [DOI] [PubMed] [Google Scholar]
- Saussez S, Cucu DR, Decaestecker C, Chevalier D, Kaltner H, André S, Wacreniez A, Toubeau G, Camby I, Gabius H-J, et al. Galectin-7 (p53-induced gene 1): A new prognostic predictor of recurrence and survival in stage IV hypopharyngeal cancer. Ann Surg Oncol. 2006;13:999–1009. doi: 10.1245/ASO.2006.08.033. doi:10.1245/ASO.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Solís D, Maté MJ, Lohr M, Ribeiro JP, López-Merino L, André S, Buzamet E, Cañada FJ, Kaltner H, Lensch M, et al. N-domain of human adhesion/growth-regulatory galectin-9: Preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution. Int J Biochem Cell Biol. 2010;42:1019–1029. doi: 10.1016/j.biocel.2010.03.007. doi:10.1016/j.biocel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Solís D, Romero A, Kaltner H, Gabius H-J, Díaz-Mauriño T. Different architecture of the combining site of the two chicken galectins revealed by chemical mapping studies with synthetic ligand derivatives. J Biol Chem. 1996;271:12744–12748. doi: 10.1074/jbc.271.22.12744. doi:10.1074/jbc.271.22.12744. [DOI] [PubMed] [Google Scholar]
- Sturm A, Lensch M, André S, Kaltner H, Wiedenmann B, Rosewicz S, Dignass AU, Gabius H-J. Human galectin-2: Novel inducer of T cell apoptosis with distinct profile of caspase activation. J Immunol. 2004;173:3825–3837. doi: 10.4049/jimmunol.173.6.3825. [DOI] [PubMed] [Google Scholar]
- Umemoto K, Leffler H, Venot A, Valafar H, Prestegard JH. Conformational differences in liganded and unliganded states of Galectin-3. Biochemistry. 2003;42:3688–3695. doi: 10.1021/bi026671m. doi:10.1021/bi026671m. [DOI] [PubMed] [Google Scholar]
- Villalobo A, Nogale-Gonzáles A, Gabius H-J. A guide to signalling pathways connecting protein-carbohydrate interaction with the emerging versatile effector functionality of mammalian lectins. Trends Glycosci Glycotechnol. 2006;18:1–37. doi:10.4052/tigg.18.1. [Google Scholar]
- Villeneuve C, Baricault L, Canelle L, Barboule N, Racca C, Monsarrat B, Magnaldo T, Larminat F. Mitochondrial proteomic approach reveals galectin-7 as a novel BCL-2 binding protein in human cells. Mol Biol Cell. 2011;22:999–1013. doi: 10.1091/mbc.E10-06-0534. doi:10.1091/mbc.E10-06-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Singh T, Liu JH, Krzeminski M, Russwurm R, Siebert HC, Bonvin AM, André S, Gabius H-J. Activity-structure correlations in divergent lectin evolution: Fine specificity of chicken galectin CG-14 and computational analysis of flexible ligand docking for CG-14 and the closely related CG-16. Glycobiology. 2007;17:165–184. doi: 10.1093/glycob/cwl062. doi:10.1093/glycob/cwl062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.