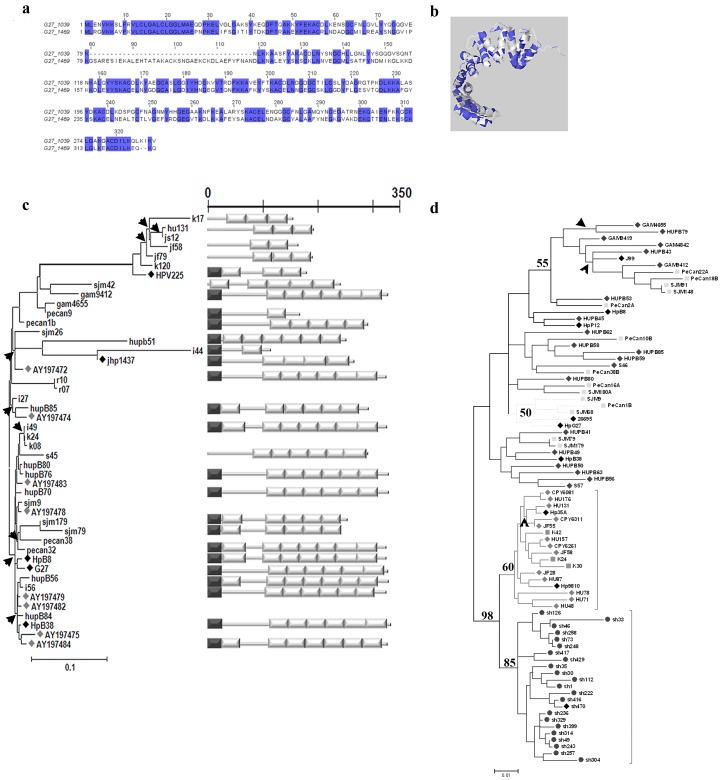
Figure 1. hcpG alleles are highly polymorphic whereas hcpC alleles are relatively conserved.
(a) Amino acid alignment of HcpC (G27_1039) and HcpG (G27_1469) from H. pylori strain G27MA. Identical amino acids are shaded in blue. (b) Mapping of amino acids shared by HcpC and HcpG onto the HcpC crystal structure. The alignment and amino acid mapping was performed using the Jalview multiple sequence editor (version 2.5.1). (c) Phylogenetic and corresponding domain architecture analyses of hcpG alleles (N = 46). Left, The ML phylogeny reconstructed using the TVM+Γ substitution model (Table S3 in File S1). Bar = 0.1 nucleotide substitutions per site and the arrowheads indicate hcpG lineages that experienced positive selection. Black diamonds, hcpG aleles obtained from sequenced H. pylori genomes; and grey diamonds, hcpG alleles obtained from GenBank. Right, domain architectures of representative hcpG alleles drawn to scale. The white rectangles are Slrs, and the black rectangles are predicted secretion signal sequences. (d) ML phylogeny of hcpC alleles (N = 81) reconstructed using the TrN+I+Γ substitution model (Table S4 in File S1). The geographic origins of the strains included are listed in Table S1 in File S1: black diamonds indicate hcpC alleles obtained from sequenced H. pylori genomes. Bar = 0.1 nucleotide substitutions per site. Arrowheads indicate hcpC lineages that experienced positive selection. Phylogenetic datasets used in generating panels (c) and (d) have been submitted to GenBank® with the following accession numbers: 1) hcpC dataset, KC007946–KC008026 and 2) hcpG dataset KC008027–KC008064).