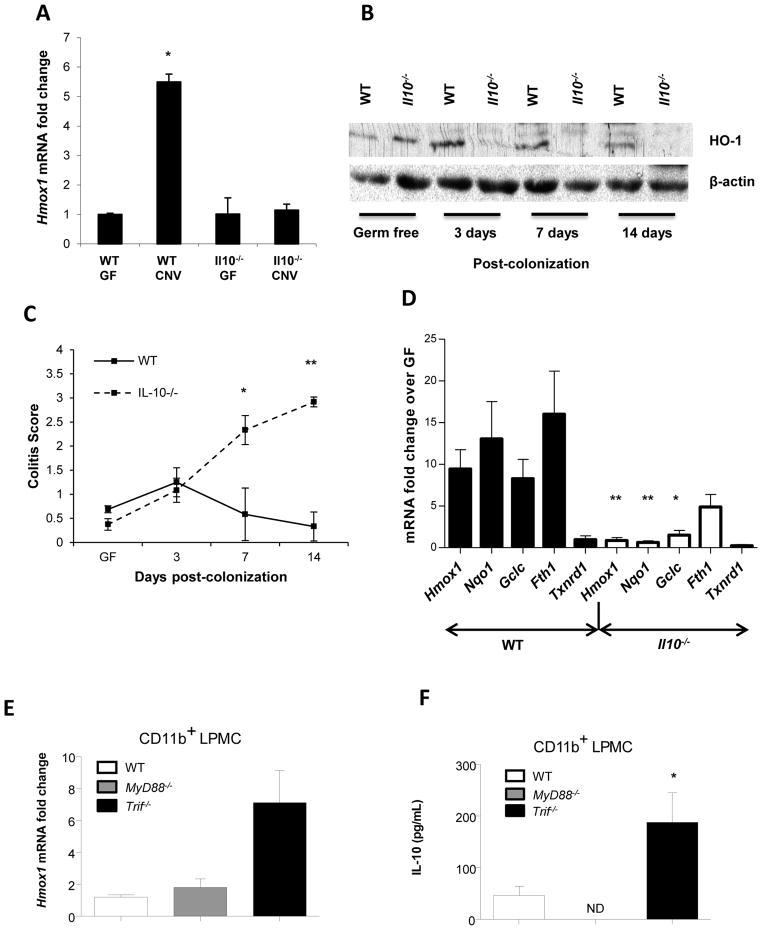
Figure 1. The enteric microbiota regulate colonic Hmox1 expression through Nrf2 and TLR dependent pathways.
WT (n = 3) and Il10−/− mice (n = 3) in GF conditions were conventionalized (CNV) with SPF enteric microbiota at 8 weeks of age. (A) Total RNA was isolated 14 days post-CNV and Hmox1 mRNA detected by real-time RT-PCR. Error bars represent mean ± SEM of three mouse colons analyzed independently. *p < .05 versus WT GF mice. (B) Total colonic protein was isolated and analyzed for HO-1 expression by Western blot. Data are representative of three independent blots on individual mouse colons. (C) Colitis scores of 8-week-old GF and CNV WT and Il10−/− mice represent mean ± SEM from three individual mouse colons. *p < .05, **p < .01 versus WT GF mice. (D) Total colonic RNA was isolated from WT (7 mice/group) and Il10−/− (7 mice/group) mice raised either GF or 14 days post-CNV. Expression of Nrf2 inducible genes (Hmox1, Nqo1, Gclc, Fth1, Txnrd1) were detected by real-time RT-PCR (normalized to β-actin mRNA). Results are from two independent experiments, presented as fold change of CNV mice relative to GF baseline controls. *p < .05, **p < .005. (E) Hmox1 expression was detected by real-time RT-PCR. Data represent mean ± SEM from 2 independent experiments with 3 mice per group, presented as fold change of CNV mice relative to GF baseline controls (normalized to β-actin mRNA). (F) Colonic CD11b+ LPMCs were isolated from WT, MyD88−/−, and Trif−/− mice. IL-10 secretion was determined by ELISA. Data represents mean ± SEM from 2 independent experiments with 3 mice per group. *p < .05.