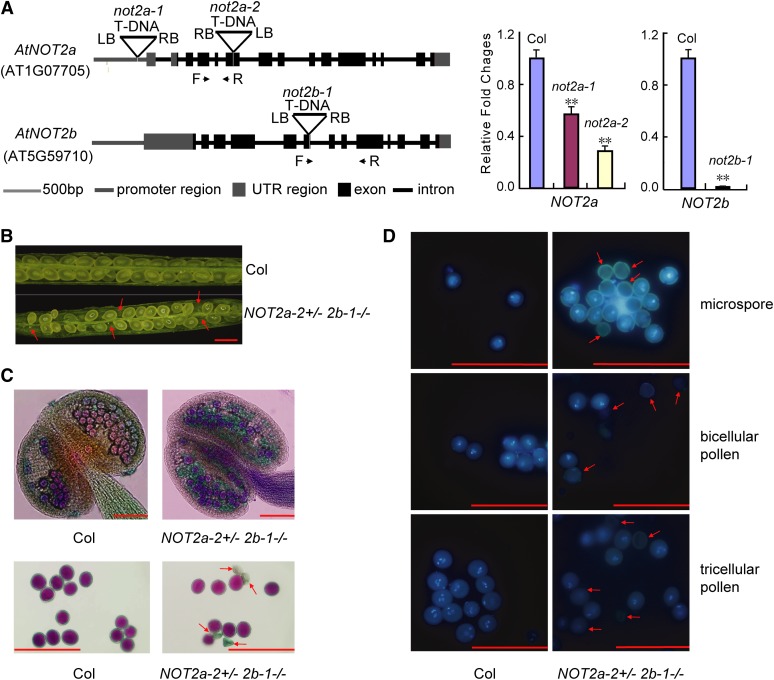
Figure 2.
Male Gametophyte Development Is Affected in Severe not2 Mutants.
(A) Diagram of NOT2a and NOT2b genes and T-DNA insertion lines. Exons and introns are shown as black boxes and lines, respectively; gray boxes and lines represent 5′or 3′ untranslated regions (UTR) and promoter regions, respectively. The T-DNA insertion sites for each mutant are indicated by triangles. Transcript levels of NOT2a and NOT2b are detected by qRT-PCR in the indicated samples (right panels). Forward (F) and reverse (R) primer locations indicated by black arrows are shown. The relative fold changes were normalized to ACTIN. Error bars represent sd. Data are given as means sd of at least three independent biological replicates. Asterisks denote Student’s t test significance for difference between the indicated samples and the wild-type control: **P < 0.01.
(B) Seed development in the wild-type Col and NOT2a-2+/− 2b-1−/− mutant siliques. Red arrows indicate aborted ovules. Bar =1 mm.
(C) Alexander’s staining of anthers and mature pollen grains in the wild type (Col) and NOT2a-2+/− 2b-1−/− mutants. Viable pollen is stained purple and degenerated pollen is green. Bars = 100 µm.
(D) DAPI staining of microspores and bicellular and tricellular pollen in the wild type (Col) and NOT2a-2+/− 2b-1−/− mutants. Red arrows indicate degenerated pollen grains. Bars = 100 µm.