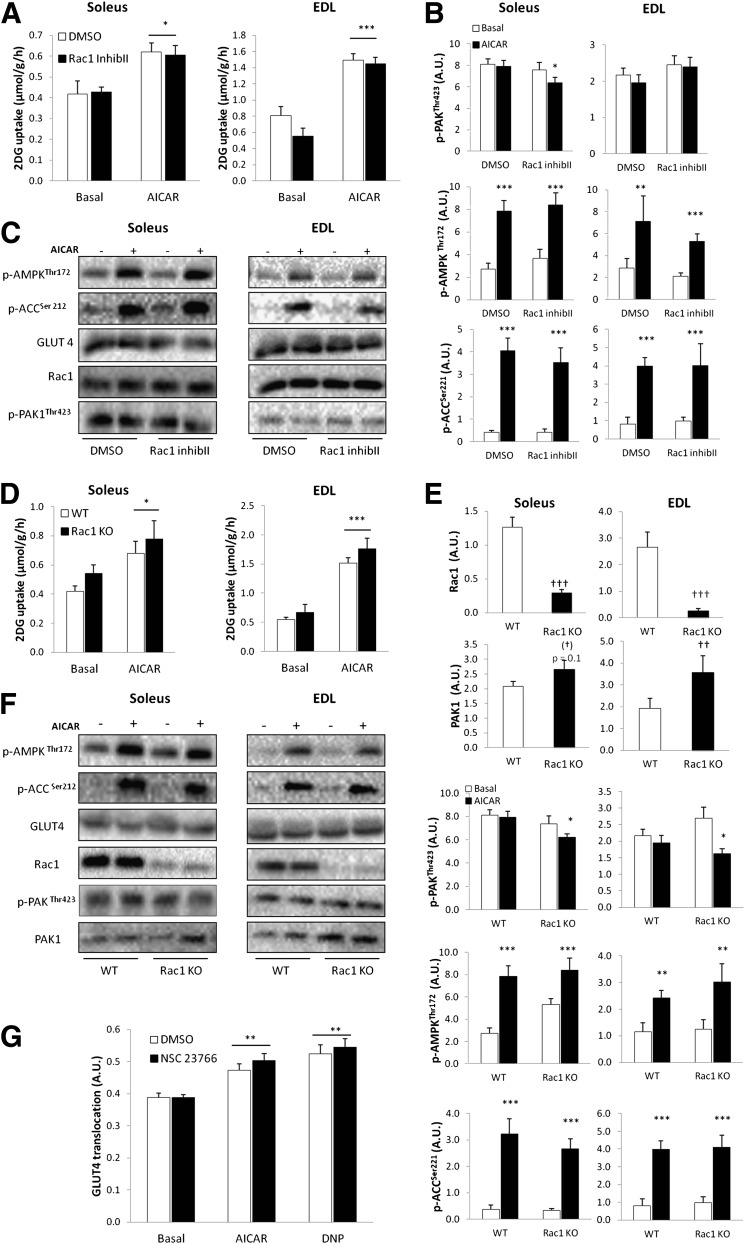
FIG. 7.
A: AICAR-stimulated (2 mmol/L) 2DG uptake in incubated isolated soleus and EDL without or with 10 μmol/L Rac1 Inhibitor II (Inhib II), 1-h preincubation (n = 8). B: Bar graphs show mean ± SEM of AICAR-induced p-AMPK Thr172, p-ACC Ser212, p-PAKThr423, in soleus and EDL without or with 10 μmol/L Rac1 Inhibitor II (Inhib II) (n = 8). C: Representative Western blots of p-AMPKThr172, p-ACCSer212, p-PAKThr423, and GLUT4 and Rac1 total proteins in response to AICAR without or with 10 μmol/L Rac1 Inhibitor II (Inhib II). D: AICAR-stimulated 2DG uptake in mouse soleus and EDL muscles from WT and muscle-specific inducible Rac1 KO (n = 8) mice. E: Bar graphs show mean ± SEM of AICAR-induced p-AMPK Thr172, p-ACC Ser212, p-PAKThr423, and total Rac1 and PAK1 protein in soleus and EDL muscles from WT or Rac1 KO mice (n = 8). F: Representative Western blots of p-AMPKThr172, p-ACCSer212, and p-PAKThr423/PAK1, and GLUT4, Rac1, and PAK1 total proteins in response to AICAR in WT and Rac1 KO soleus and EDL muscles. G: AICAR-stimulated GLUT4 translocation in C2C12-GLUT4myc myotubes in response to DNP (0.5 mmol/L) or AICAR (2 mmol/L) without or with 200 μmol/L NSC23766, 1-h preincubation (n = 6). Statistical significances between basal and AICAR are indicated by *P < 0.05; **P < 0.01; ***P < 0.001. Significant differences in AICAR-stimulated signaling are indicated by #P < 0.05. Main effect of genotype is indicated by ††P < 0.01 and †††P <0.0001. Values represent mean ± SEM.