Recent work from the Leigh lab, published in Science,1 describes the design, synthesis and operation of an artificial small-molecule capable of synthesizing peptides in a sequence-specific manner. The design of this artificial molecular machine was, in the author’s own words, “inspired by the ribosome”2; with several elements analogous to ribosomal protein synthesis (Fig. 1). The tRNA’s of conventional translation are substituted by amino acids attached to a track (or axle) by weak phenolic ester linkages. The sequence and spacing of the axle-tethered amino acids enables sequence specific peptide synthesis (analogous to the mRNA template in nature). Using copper ions to direct the process, attachment of the artificial molecular machine to the axle is mediated by a macrocycle—a large nanometer-sized molecular ring that straddles the axle and moves along its axis in a manner reminiscent of the ribosome subunits clamping to and moving along the mRNA strand. Attached to the ring is a reactive arm (a cysteine derivative bearing a trityl (Trt)-protected thiol group) which detaches the amino acids from the axle, passing them to the elongation site (a tert-butoxycarbonyl carbamate (Boc)—protected amine at the end of a glycylglycine residue), where the resulting peptide oligomer is synthesized in a single specific sequence, through native chemical elongation.3
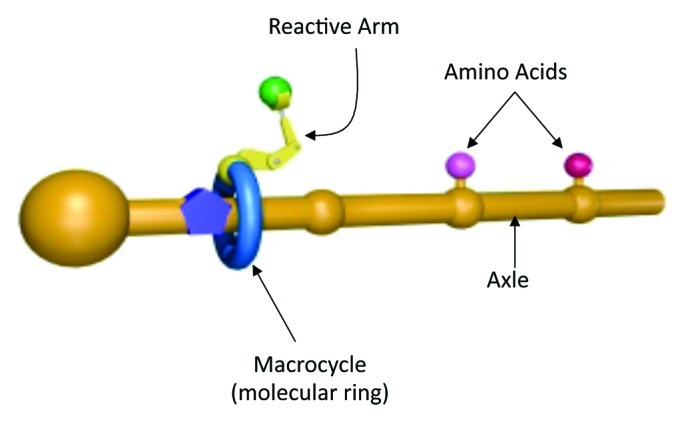
Figure 1. Schematic representation of the synthetic ribosome; illustrating the macrocycle with attached reactive arm and the axle with attached amino acids
The synthesis reaction is initiated by acid-catalyzed cleavage of the Boc and trityl protecting groups; activating the molecular machine, which then operates at 60°C under microwave heating in the presence of a reducing agent (which cleaves any disulphide bonds formed through thiol oxidation). Once the thiolate residue of the catalytic cysteine group is deprotected, it undergoes a transacylation reaction with the first amino acid phenolic ester that blocks the ring as it slides along the axle. The subsequently formed phenylalanine thioester then transfers the amino acid to the glycylglycine amine group. This sequence simultaneously transfers the amino acid to the end of the growing peptide and regenerates the catalytic thiolate group, ready for cleavage and transfer of further amino acids. Once the covalent bond linking the amino acid to the axle is broken, the macrocycle is free to move further along the axle until it reaches the next amino acid in the sequence, and the entire process is repeated. When the last amino acid on the axle is cleaved the macrocycle detaches with the newly formed, full length peptide attached. Unlike ribosomal translation, the artificial molecular machine synthesizes the peptide in the opposite direction i.e., from the C- to the N-terminus.
Proof that the process works was obtained using a combination of HPLC (which showed that none of the starting material remained), 1H NMR and mass spectrometry—which identified the completely deacylated thread (or axle) and the macrocycle bearing the synthesized hexapeptide (Piv)AlaLeuPheGlyGlyCys [Piv,COC(CH3)3]. The peptide sequence was confirmed by tandem mass spectrometry (MS/MS).
Although only about one-tenth the size of nature’s ribosomes,4 Leighs’s peptide synthesizer is capable of producing peptides in a scale of tens of milligrams (corresponding to parallel synthesis by ~108 molecular machines). However, taking ~12 h to make each peptide bond, the synthetic system remains a “primitive analog of the ribosome”2 which is capable of synthesizing ~15 to 20 amide bonds per second. Furthermore, the rotaxane destroys the code it reads, and can produce only relatively short peptide strands. Nonetheless, the current study provides a valuable proof of concept; a first generation prototype which is likely to be improved upon. One suggested upgrade is to replace the preloaded axle with amino acid binding sites (as opposed to the amino acids themselves). Using this approach, once the amino acids have been picked in the first round, the axle could reload (by binding fresh amino acids from the bathing solution), thereby reversing the loss of sequence information on the strand.
Leigh’s rotaxane is the latest in a growing list of artificial molecular machines inspired by biology and with diverse functions and applications, including information storage,5 targeted drug delivery,6 mechanical work7 and even the ability to switch “on” and “off” catalytic activity.8 Such naturally inspired molecular machines represent a new and exciting branch of biomolecular engineering—in the words of Leigh himself “that’s how biology does it, so why can’t we?”2.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/23640
References
- 1.Lewandowski B, De Bo G, Ward JW, Papmeyer M, Kuschel S, Aldegunde MJ, et al. Sequence-specific peptide synthesis by an artificial small-molecule machine. Science. 2013;339:189–93. doi: 10.1126/science.1229753. [DOI] [PubMed] [Google Scholar]
- 2.Peplow M. Molecular robot mimics life's protein-builder. Nature 2013; [modified 2013 Jan 10; cited 2013 Jan 15]. Available from: http://www.nature.com/news/molecular-robot-mimics-life-s-protein-builder-1.12190
- 3.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–9. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 4.Ramakrishnan V. Unraveling the structure of the ribosome (Nobel Lecture) Angew Chem Int Ed Engl. 2010;49:4355–80. doi: 10.1002/anie.201001436. [DOI] [PubMed] [Google Scholar]
- 5.Green JE, Choi JW, Boukai A, Bunimovich Y, Johnston-Halperin E, DeIonno E, et al. A 160-kilobit molecular electronic memory patterned at 10(11) bits per square centimetre. Nature. 2007;445:414–7. doi: 10.1038/nature05462. [DOI] [PubMed] [Google Scholar]
- 6.Dam HH, Caruso F. Modular click assembly of degradable capsules using polyrotaxanes. ACS Nano. 2012;6:4686–93. doi: 10.1021/nn301045z. [DOI] [PubMed] [Google Scholar]
- 7.Berná J, Leigh DA, Lubomska M, Mendoza SM, Pérez EM, Rudolf P, et al. Macroscopic transport by synthetic molecular machines. Nat Mater. 2005;4:704–10. doi: 10.1038/nmat1455. [DOI] [PubMed] [Google Scholar]
- 8.Blanco V, Carlone A, Hänni KD, Leigh DA, Lewandowski B. A rotaxane-based switchable organocatalyst. Angew Chem Int Ed Engl. 2012;51:5166–9. doi: 10.1002/anie.201201364. [DOI] [PubMed] [Google Scholar]


