Figure 1.
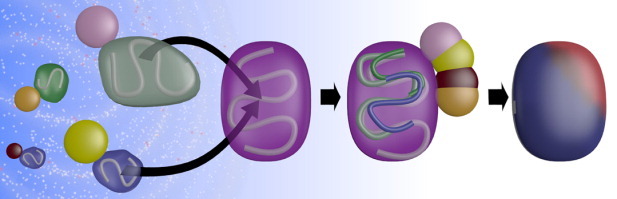
Exploring the protein universe using structural BLAST. To infer function of a given query protein (purple), the universe of structures (left) is searched broadly for proteins that are structurally similar to the query and each one is placed in the query coordinate system (curved arrows) by superposing the protein backbones (shown schematically within the surface of each protein). In the specific application shown in the figure, binding partners (small molecules, other proteins, nucleic acids, etc.) of the structural neighbors (shown as pink, yellow, magenta, and orange circles) are also transformed into the coordinate system of the query (middle panel). The query is then analyzed to find surface residues that show a statistical propensity to contact ligands from structural neighbors. This information is represented as a heat map on the surface of the query (right panel). Regions where the query is likely to bind other molecules are shown in red and unlikely regions are shown in blue.
