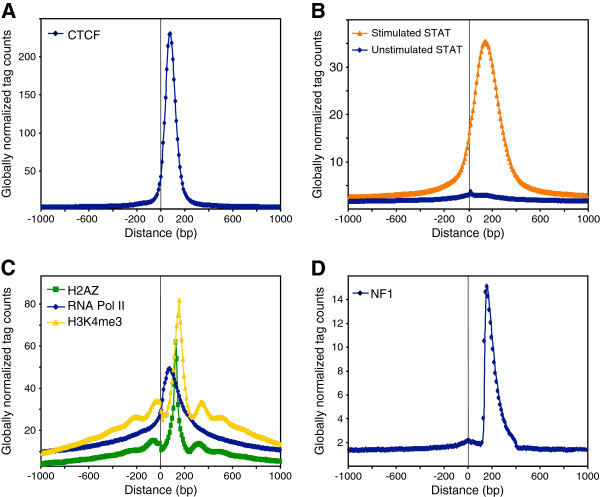
Figure 2.
Positional correlation of tags mapping on plus and minus strand from other ChIP-Seq experiments. Co-localized tag counts greater than 10 were set to 10 and tags overlapping repeated DNA sequences, as defined by RepeatMasker, were filtered out. Tags mapping on the plus strand were centered to position 0 and the distribution of tags mapping on the minus strand is displayed as a function of the distance to the centered plus strand tags, using ChIP-Seq datasets generated for CTCF (panel A), STAT1 from stimulated and unstimulated cells (B), the H2AZ histone variant, RNA Pol II, and histone H3 tri-methylated on lysine 4 (H3K4me3) (C), or NFI (D).