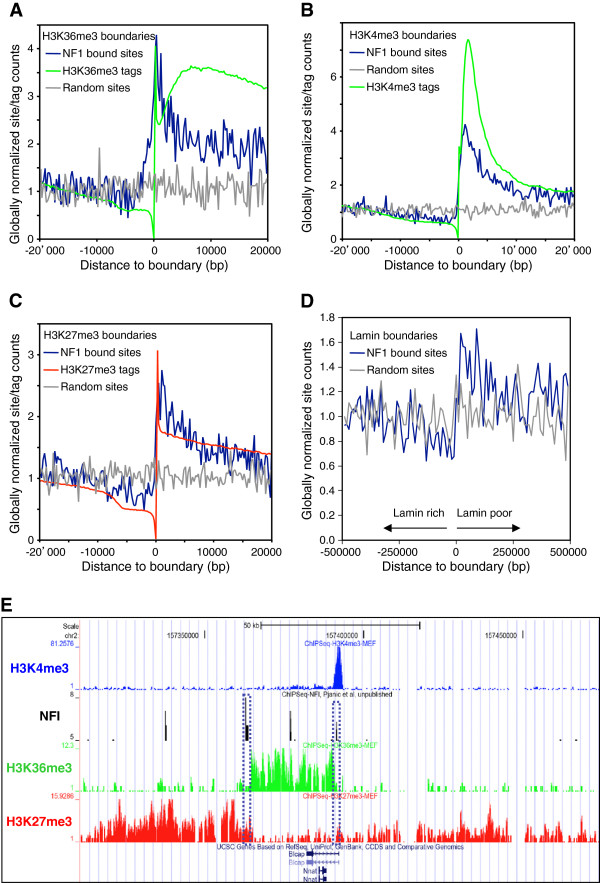
Figure 4.
NFI binding is associated with chromatin and nuclear lamina boundaries. The mouse genome was partitioned into histone modification-poor and enriched segments, and the genomic positions of boundaries featuring transitions from modification-poor to enriched regions were centered and aligned at position 0 of the X-axis, from (A) 14,632 H3K36me3 boundaries, (B) 31,662 H3K4me3 boundaries (C) 29,122 H3K27me3 modification boundaries. The distribution of 4,794 in vivo NFI sites was compared with the distribution of 4,794 randomly chosen genomic positions sampled from each chromosome according to the number of occupied NFI sites. The average numbers of NFI or random sites were normalized globally with the genome-wide average number of sites in 250 bp window. The distribution of NFI and random sites was also aligned to the anchor spots of DNA to the nuclear lamina using 2 kb windows (D), where the arrows indicate the poor and enriched genomic regions for the nuclear lamina lamin B component. (E) Example of NFI in vivo binding sites located at the boundaries of open and close chromatin markers, H3K36me3 and H3K27me3, in mouse embryonic fibroblasts. NFI binding sites define the region of open chromatin surrounding the Blcap and Nnat gene locus. NFI sites at the boundary positions are enclosed with a dotted line.