Figure 6.
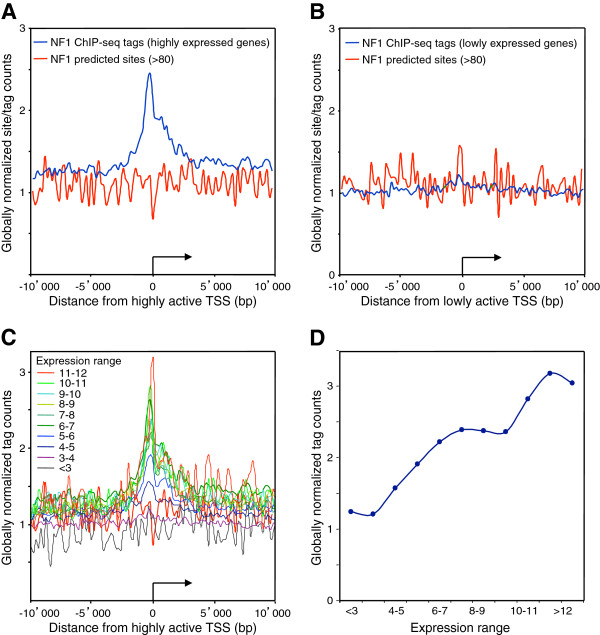
NFI tag count is increased near transcription start sites of highly-expressed genes. Average NFI ChIP-Seq tag counts were calculated in windows of 200 bp for regions 10 kb up- and down- stream of the RefSeq annotated transcription start sites (TSS). TSS were oriented and the broken arrows indicate the initiation site and the direction of transcription. Tag counts were normalized globally, as a fold increase over the genome average tag count in a window of 200 bp. NFI predicted sites were mapped on the reference mouse genome mm9 with a matrix cut-off score 67. The NFI predicted site count was normalized in the same way, as a fold increase over the average site count in a bin window of 200 bp. (A) Highly-expressed genes with Affymetrix expression levels greater than 8. (B) Low- or non- expressed genes with Affymetrix expression levels lower than 4. Genes were separated into 11 groups according to their Affymetrix expression level (<3, 3–4, 4–5, 5–6, 6–7, 7–8, 8–9, 9–10, 10–11, 11–12, >12). (C) For each of these groups, the average NFI ChIP-Seq tag counts was calculated in windows of 200 bp for regions 10 kb up- and down- stream of the RefSeq annotated transcription start sites (TSS). Tag counts were normalized globally, as a fold increase over the genome average tag count in a window of 200 bp. (D) Peak maxima for each of the gene groups found at the TSS was plotted against the expression level.