Abstract
Interleukin-2 is a pleiotropic cytokine produced after antigen activation that plays pivotal roles in the immune response. Discovered as a T-cell growth factor, IL-2 additionally promotes CD8+ T cell and NK cell cytolytic activity, and modulates T cell differentiation programs in response to antigen, promoting naïve CD4+ T cell differentiation into T helper-1 (Th1) and T helper-2 (Th2) cells while inhibiting T helper-17 (Th17) and T follicular helper (Tfh) cell differentiation. Moreover, IL-2 is essential for the development and maintenance of T regulatory (Treg) cells and for activation-induced cell death, thereby mediating tolerance and limiting inappropriate immune reactions. In this review, we focus on the molecular mechanisms and complex cellular actions of IL-2, its cooperative and opposing effects with other cytokines, and how both promoting and blocking the actions of IL-2 are being utilized in clinical medicine.
Introduction
Interleukin-2 (IL-2) was first discovered over 35 years ago as an activity present in supernatants of activated human T cells that mediates T cell growth and proliferation (Morgan et al., 1976); previously reviewed in (Boyman and Sprent, 2012; Kim et al., 2006; Lin and Leonard, 2000; Malek and Castro, 2010). This four α-helix bundle type 1 cytokine (Bazan, 1990) was the first type 1 cytokine cloned (Taniguchi et al., 1983) and the first type 1 cytokine for which a receptor component was cloned (Leonard et al., 1984; Nikaido et al., 1984) and has served as a paradigm for other cytokines, particularly because it is one of two cytokines to share the IL-2 receptor β chain (IL-2Rβ) and one of six cytokines to share the common cytokine receptor γ chain, γc (Figure 1), with both of IL-2Rβ and γc having been discovered as components of the IL-2 receptor (Leonard, 2001).
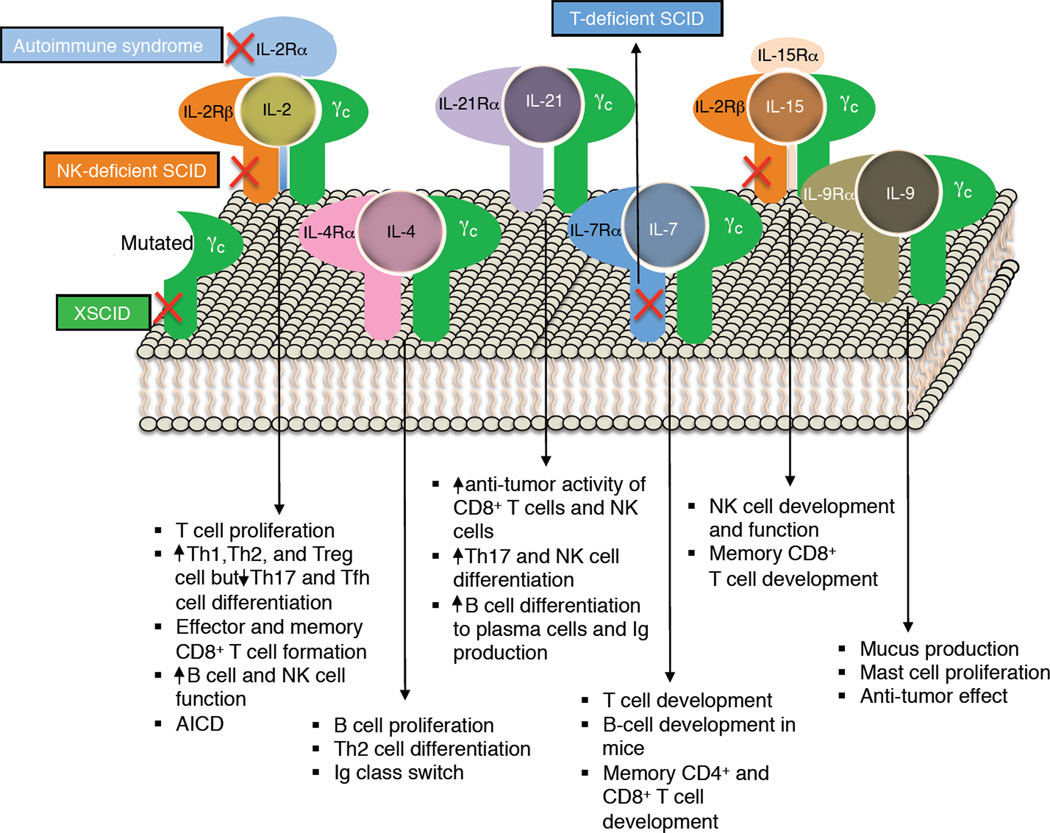
Figure 1. The γc family of cytokines.
Shown are the receptors for IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21, as well as major actions for these cytokines. Crosses in red indicate that mutation of IL2RG gene, which encodes γc, results in X-linked severe combined immunodeficiency in humans (XSCID, where both T cells and NK cells are greatly diminished [T−B+NK− SCID]), mutation of IL2RA results in an autoimmune syndrome, defective expression of IL2RB results in NK-deficient SCID (where T and B cells remain [T+B+NK− SCID]), and mutation of IL7R causes T-cell selective form of SCID, where B and NK cell numbers are normal (T−B+NK+ SCID). JAK3 is not shown as it interacts with the cytoplasmic domain of γc; however, mutations in JAK3, as noted in the text, cause a T−B+NK− for SCID, like XSCID.
Besides its potent T-cell growth factor activity, IL-2 induces proliferation of natural killer (NK) cells and augments their cytolytic activity as well as that of lymphokine-activated killer cells (Siegel et al., 1987), promotes antibody production and proliferation by B cells (Mingari et al., 1984), and is essential for activation-induced cell death (AICD), which is important for homeostasis and the elimination of potentially harmful auto-reactive cells, at least in part by a Fas and FasL-dependent mechanism (Lenardo et al., 1999). IL-2 also drives the development of CD4+FOXP3+ regulatory T cells (Treg cells), which have suppressor function and mediate tolerance (Littman and Rudensky, 2010; Sakaguchi et al., 2008; Shevach, 2009). More recently, the range of recognized actions of IL-2 has expanded, with roles in promoting the differentiation of T helper 1 (Th1) (Liao et al., 2011; Shi et al., 2008) and Th2 cells (Cote-Sierra et al., 2004; Liao et al., 2008), while inhibiting Th17 (Laurence et al., 2007) and T follicular helper (Tfh) cell (Ballesteros-Tato et al., 2012) development, but nevertheless promoting Th17 cell expansion once cells develop (Amadi-Obi et al., 2007). IL-2 also is critical for production of IL-9 (Schmitt et al., 1994). Thus, IL-2 has broad essential biological actions, not only driving T cell proliferation and modulating effector cell differentiation, but also limiting potentially dangerous autoimmune reactions. Herein, we discuss the molecular and cellular biology of IL-2, its signaling mechanism, and actions, as well as its relationship with the five other cytokines (IL-4, IL-7, IL-9, IL-15, and IL-21) that share components of the IL-2 receptor. Finally, we discuss the use of IL-2 as a therapeutic agent and the utility of blocking the action of IL-2 and related cytokines using Janus kinase (JAK) inhibitors, an exciting new class of immunosuppressive drugs.
IL-2
IL-2 is a 15.5 kDa type 1 four α-helical bundle cytokine produced primarily by CD4+ T cells following antigen stimulation but also produced to a lesser extent by CD8+ cells (Paliard et al., 1988), NKT cells (Yui et al., 2004), activated dendritic cells (DCs) (Granucci et al., 2001), and mast cells (Hershko et al., 2011). In T cells, induction of IL-2 transcription requires two signals, mediated by calcium and protein kinase C. IL-2 transcription is mediated by multiple transcription factors (Figure 2A), including nuclear factor of activated T cells (NFAT) family proteins (Muller and Rao, 2010), activator protein-1 (AP-1, FOS-JUN family dimers), nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), and the octamer transcription factor, OCT-1 (Kim et al., 2006). Of five NFAT proteins (NFATC1, NFATC2, NFATC3, NFATC4, and NFAT5), all but NFAT5 are expressed in the cytosol, with NFATC1 and NFATC2 highly expressed in lymphocytes (Muller and Rao, 2010) and mice lacking both of these proteins express essentially no T cell receptor (TCR)-induced IL-2 (Peng et al., 2001). NFAT translocation to the nucleus is blocked by cyclosporine A and FK-506 (Flanagan et al., 1991). TCR engagement activates AP-1, which with NFAT, binds to composite sites in the IL-2 promoter (Figure 2A) (Muller and Rao, 2010). In anergic cells, the distal AP-1 site binds the transcription factors CREB and/or CREM instead of AP-1; CREB activates expression of the transcription factors cJUN, cFOS, FRA2 and FOSB, and a dominant negative CREB greatly decreases TCR-induced IL-2 expression (Kim et al., 2006). FOXP2 also cooperates with NFAT to drive IL-2 expression, whereas FOXP3 can replace FOXP2 to inhibit IL-2 expression while inducing the Treg cell markers, IL-2Rα and CTLA4 (Wu et al., 2006). NF-κB binds to two sites in the IL-2 promoter, including one in the CD28 response element, and expression of IL-2 is elevated in Nfkb1−/− mice (Kim et al., 2006), supporting the model that p50 homodimers repress IL-2 expression, whereas complexes containing p65 or c-Rel activate its expression. Oct1 and Oct2 bind to octamer binding sites and cooperate with AP-1 for IL-2 gene induction by phorbol myristate acetate (PMA) + ionomycin; The transcription factors SP1, Egr1, and GABP also act as positive regulators (Kim et al., 2006). TRIM28, a component of heterochromatin, is phosphorylated after TCR stimulation and promotes IL-2 expression, with diminished IL-2 production in mice in which the Trim28 gene was conditionally deleted as well as in Trim28-siRNA treated human Jurkat T cells (Chikuma et al., 2012).
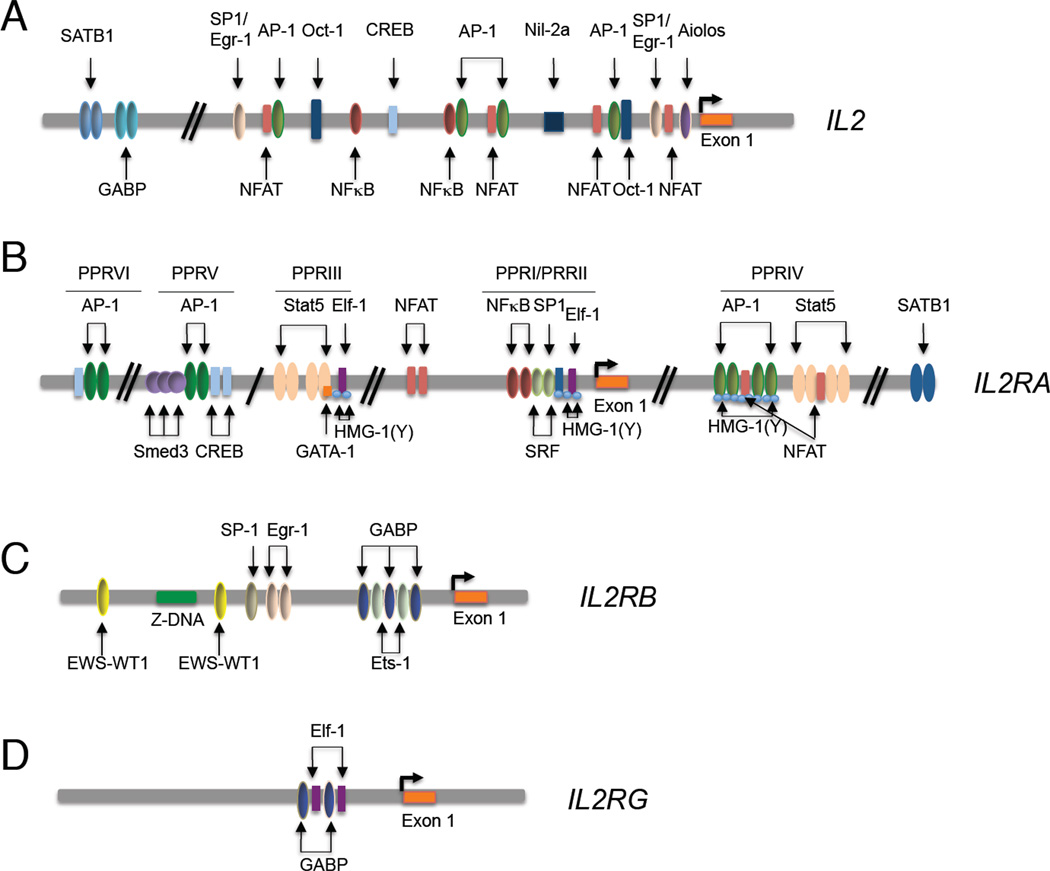
Figure 2. Factors regulating the IL2 (A), IL2RA (B), IL2RB (C), and IL2RG (D) genes.
For each gene, the binding locations of transcription factors are shown. For some of these factors, there are only in vitro data that indicate their importance, whereas for others such as STAT5A and STAT5B, extensive in vivo data have established their importance (e.g., of STAT5A and STAT5B for regulation of IL-2Rα expression). (A) Multiple factors, including for example NFAT, AP1, and NF-κB bind to and regulate the IL2 gene. (B) In the IL2RA gene, PRRI binds SP1, SRF, and NF-κB; PRRII binds Elf-1 as well as HMG-I and/or HMG-Y; PRRIII binds STAT5A, STAT5B, ELF1, and GABP, as well as HMG-I and/or HMG-Y, PRRIV binds NFAT, AP1, STAT5A, and STAT5B; PRRV binds SMAD3, AP1, and CREB-ATF factors, and PRRVI binds AP1 and CREB-ATF factors. (C) Factors including ETS1 bind to and regulate the IL2B gene. (D) Only limited information is available regarding the factors regulating IL2RG.
IL-2 transcription also is negatively regulated. NIL2A (TCF-8) is encoded by ZEB1, a zinc finger E box binding protein that binds ~105 bp 5’ of the transcription start site and suppresses transcription (Williams et al., 1991). Prior to activation, the T-lineage factor SATB1 binds to the human IL-2 promoter and in a phosphorylation-dependent manner recruits histone deacetylase-1 (HDAC1) and silences Il2 expression, whereas after HIV-1 infection, binding of HIV Tat as well as CBP and/or p300 to HDAC1 induces IL-2 (Pavan Kumar et al., 2006). BLIMP-1 and Aiolos (encoded by Prdm1 and Ikzf3, respectively) also repress IL-2 expression. IL-2 induces BLIMP-1, which binds IL-2 and cFos promoter regions and inhibits their expression (Martins et al., 2008). In Th17 cells, Aiolos is induced in a signal transducer and activator of transcription-3 (STAT3)- and aryl hydrocarbon receptor (Ahr)-dependent fashion and binds a CCTCCCATGC motif in the Il2 promoter in Th17 but not Th1 or Th0 cells, suppressing Il2 expression in Th17 cells (Quintana et al., 2012). Interestingly, microRNAs (miRNAs) also play a role in regulation of IL-2 expression. For example, miR146a, which is induced by TCR stimulation in primary T cells, can impair AP-1 production and IL-2 expression (Curtale et al., 2010), whereas miR9 induced by TCR stimulation enhances IL-2 expression, at least in part, by suppressing PRDM1 expression (Thiele et al., 2012), and Mir184 represses expression of IL-2 in umbilical cord CD4+ T cells (Weitzel et al., 2009)
Three classes of IL-2 receptors
IL-2 signals via specific receptors (Robb et al., 1981), with three classes of cell surface receptors formed by various combinations of three IL-2 receptor (IL-2R) subunits (Figure 3), IL-2Rα, IL-2Rβ, and IL-2Rγ (Kim et al., 2006; Lin and Leonard, 2000; Malek and Castro, 2010). IL-2Rα (CD25) was originally called Tac antigen based on the demonstration that anti-Tac monoclonal antibody (mAb) (Uchiyama et al., 1981) blocked the binding of IL-2 (Leonard et al., 1982). IL-2Rβ (CD122) (Sharon et al., 1986; Teshigawara et al., 1987; Tsudo et al., 1986) is also part of the IL-15R complex (Giri et al., 1994), and IL-2Rγ (CD132) (Takeshita et al., 1992) was renamed as the common cytokine receptor γ chain, γc (Leonard et al., 1995; Noguchi et al., 1993a; Russell et al., 1993); γc is now known to be shared by IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 (Leonard, 2001; Rochman et al., 2009).
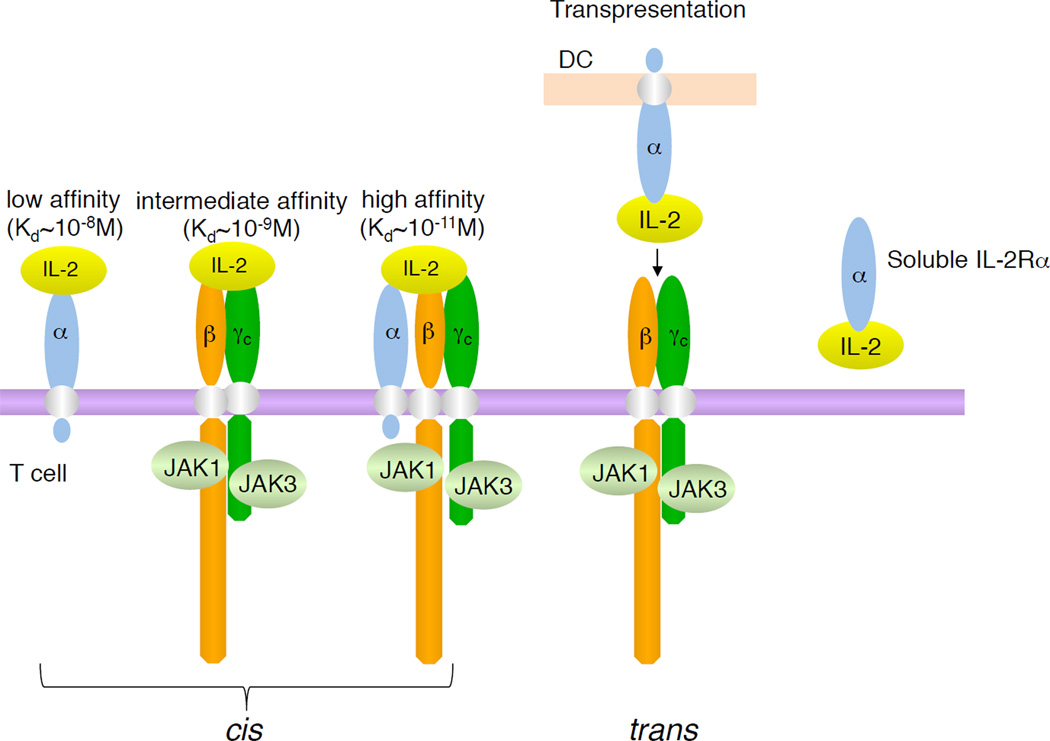
Figure 3. Classes of IL-2 receptors.
Shown are the three classes of IL-2 receptors (high-affinity, intermediate affinity, and low affinity), with receptor composition, Kd’s, receptor composition, and associated JAK kinases. Also shown is trans-presentation of IL-2 by a DC that expresses IL-2Rα to a T cell that expresses IL-2Rβ and γc. On the right is the soluble IL-2 receptor (soluble IL-2Rα) with bound IL-2. As discussed in the text, IL-2 is produced primarily by activated CD4+ T cells.
IL-2Rα is absent or minimally expressed on resting T and NK cells but its transcription is potently induced on T cells stimulated via the TCR or IL-2 (Depper et al., 1985; Depper et al., 1984; Leonard et al., 1985) or on NK cells stimulated with IL-2 (Siegel et al., 1987); the expression level of the IL-2Rβ chain is low on T cells but is induced by certain stimuli, including IL-2 (Siegel et al., 1987); γc is also expressed on these cells but is less inducible than IL-2Rα or IL-2Rβ (Cao et al., 1993). IL-2Rα is the “low-affinity” IL-2 receptor (Kd ~10−8 M); IL-2Rβ binds poorly (Kd ~ 1 µM) by itself, but the combination of IL-2Rβ with γc forms the intermediate-affinity IL-2 receptor (Kd ~10−9 M); and all three subunits together form the high-affinity IL-2 receptor (Kd ~ 10−11 M) (Takeshita et al., 1992), the structure for which has been solved (Stauber et al., 2006; Wang et al., 2005). High affinity IL-2 binding results from IL-2’s rapid on and off rates to IL-2Rα coupled to slower on and off rates to intermediate affinity receptors (Lowenthal and Greene, 1987; Wang and Smith, 1987). When IL-2Rα and IL-2Rβ are co-expressed without γc, “pseudo-high affinity” binding (Kd ~ 10−10 M)(Arima et al., 1992) is achieved but no signaling occurs. The intermediate and high affinity receptors are functional, corresponding to the requirement for heterodimerization of IL-2Rβ and γc cytoplasmic domains for signaling (Nakamura et al., 1994; Nelson et al., 1994). Intermediate affinity receptors are present on resting T (Zhang et al., 1998) and NK cells (Siegel et al., 1987), whereas high-affinity receptors are expressed by activated lymphocytes (Robb et al., 1981). Binding of IL-2 to intermediate affinity receptors induces cell growth and cytolytic activity (Siegel et al., 1987) and IL-2Rα transcription (Depper et al., 1985). After T-cell activation, IL-2Rα is rapidly induced and high-affinity receptors form, increasing responsiveness to IL-2. Although IL-2 primarily acts as a soluble factor via intermediate and high-affinity receptors, like IL-15, IL-2 can be presented in trans, where IL-2 bound to IL-2Rα on one cell stimulates another cell that expresses IL-2Rβ and γc (Wuest et al., 2011)(Figure 3). However, unlike the relatively high-affinity binding of IL-15 to IL-15Rα (Waldmann, 2006), IL-2 binds with relatively low affinity to IL-2Rα with rapid on and off rates, as noted above. Accordingly, cis-based IL-2 signaling is likely favored whenever a cell expresses all three chains, and trans-signaling may require relatively high local concentrations of IL-2. In addition to cell surface IL-2Rα, IL-2Rα can exist in a soluble form (sIL-2Rα) that can be released from the cell surface (Figure 3), including in infectious disorders, transplantation rejection, and autoimmune inflammatory states, with an elevated amount of sIL-2Rα being detected in certain hematologic malignancies. In some diseases, the concentration of sIL-2Rα correlates with disease activity and prognosis (Rubin and Nelson, 1990).
Interestingly, IL-2Rα is expressed on a fraction of CD4−CD8− thymocytes (Godfrey et al., 1993), but such IL-2Rα expression may reflect the activation state or ability to trans-present IL-2 rather than responsiveness to IL-2. IL-2Rα is also expressed by DC populations (Driesen et al., 2008), for example after CpG or CD40 ligand stimulation of human plasmacytoid DCs (Naranjo-Gomez et al., 2007). Both cell surface and soluble IL-2Rα are expressed by primary BDCA-1+ myeloid DCs stimulated with tumor necrosis factor –α (TNF-α) and prostaglandin E2, as well as on tumor-associated DCs. These cells might sequester IL-2 and diminish T-cell proliferation, thus contributing to immunosuppression by DCs (von Bergwelt-Baildon et al., 2006), but IL-2Rα-expressing DCs can also trans-present IL-2, with agonistic effects, consistent with inhibition of T cell activation by antigen-specific DCs and diminished brain inflammation in multiple sclerosis patients treated with daclizumab, a humanized antibody to IL-2Rα (Wuest et al., 2011).
Regulation of IL-2 receptor expression
The three IL-2R chains are independently regulated. IL-2Rα is transcriptionally induced by stimulation via the T-cell receptor (Leonard et al., 1985), by cytokines including IL-1, IL-2, IL-7, IL-12, IL-15, TNF-α, and transforming growth factor-β (TGF-β), by transactivator proteins Tax of human T cell leukemia virus-1 (HTLV-1) and TaxII of HTLV-2, and by activators of protein kinase C (Kim et al., 2006). Of six positive regulatory regions (PRRI, PRRII, PRRIII, PRRIV, PRRV, and PRRVI) in the IL-2Rα gene (Figure 2B) (Kim et al., 2006), all but PRRIII contribute to antigen and mitogenic stimulation. PRRIII, PRRIV, and additional regions of the IL-2Rα gene based on chromatin immunoprecipitation-Sequencing (ChIP-Seq) data (Lin et al., 2012) are IL-2-response elements that contain interferon-γ activation sequence (GAS) motifs and bind STAT5 proteins. Correspondingly, IL-2-induced IL-2Rα expression is diminished in Stat5a−/− and Stat5b−/− mice (Imada et al., 1998; Nakajima et al., 1997). Whereas IL-2-mediated induction of the IL2RA gene is dependent on STAT proteins, its induction by TCR, TNF-α, and Tax proteins of HTLV-1 or HTLV-2, requires NF-κB binding (Kim et al., 2006) (Figure 2B). NFAT sites are also present, consistent with diminished IL-2Rα expression in Nfatc2−/− mice (Schuh et al., 1998). TGF-β cooperates with TCR signaling to act via PRRV, which has a SMAD (“small mothers against decpentaplegic”) binding site (Kim et al., 2005). Like the Il2 gene, SATB1 (special AT-rich sequencing-binding protein-1) binds to and suppresses the Il2ra gene, with elevated Il2ra expression in Satb1−/− double positive (DP) thymocytes (Alvarez et al., 2000). Two negative regulatory elements, NRE-1 and NRE-2, are also present in the gene (Kim et al., 2006).
IL-2Rβ is expressed by multiple lympho-hematopoietic populations of cells, including NK cells, resting T cells, monocytes, and neutrophils; on T cells, TCR stimulation, PMA, IL-2, and IL-4 each augments IL-2Rβ expression via both transcriptional and post-transcriptional regulation (Kim et al., 2006). The IL-2Rβ promoter binds the Ets family proteins, Ets1 and GABP, as well as Egr1 and Sp1 (Lin et al., 1993; Lin and Leonard, 1997)(Figure 2C); Ets1 binds to the IL-2Rβ promoter, with decreased IL-2Rβ expression in Ets1−/− NK cells (Ramirez et al., 2012). The Ewing sarcoma gene-Wilms tumor suppressor (EWS-WT1) fusion transcription factor also binds to and augments IL-2Rβ expression (Wong et al., 2002). Interestingly, there is a run of Z-DNA within this gene, but its physiological significance is unknown (Kim et al., 2006). IL-2Rβ and IL-2Rα expression was reported on fibroblasts (Gruss et al., 1996), but the significance of such expression also is unclear.
Like IL-2Rβ, IL-2Rγ is constitutively expressed and mainly restricted to lymphohematopoietic cells (Cao et al., 1993). IL-2 and IFN-γ are each shown to enhance and TGF-β1 to inhibit IL-2Rγ expression in human monocytes (Bosco et al., 1994), but little is still known about IL2RG regulation (Figure 2D).
Signaling via the IL-2 receptor
After IL-2 engagement, the IL-2 receptor complex is rapidly internalized, with IL-2Rα located in early transferrin+ endosomes that recycle to the plasma membrane, whereas IL-2Rβ and γc are targeted to Rab7+ vesicles that are sorted towards degradation (Hemar et al., 1995). Correspondingly, IL-2Rα is much more stable on the cell surface than are IL-2Rβ and γc. Following receptor binding, IL-2 activates multiple signaling pathways (Figure 4). Heterodimerization of the IL-2Rβ and γc cytoplasmic domains leads to activation of Janus family tyrosine kinases, JAK1 and JAK3, with JAK1 associating with IL-2Rβ and JAK3 with γc (Boussiotis et al., 1994; Miyazaki et al., 1994; Russell et al., 1994). The JAK kinases activate each other and phosphorylate key residues in IL-2Rβ. Phosphorylation of Y338 in humans (Y341 in mice) allows association of the SHC adaptor protein (Friedmann et al., 1996), which provides a platform for Ras-MAP kinase activation and promotes cell growth. Phosphorylation of Y392 and Y510 in humans (Y395 and Y498 in mice) mediates recruitment of STAT1, STAT3, STAT5A and STAT5B, with most potent and sustained activation of STAT5 proteins (Friedmann et al., 1996; Lin et al., 2012). IL-2 also activates the phosphoinositol 3-kinase (PI 3-kinase)-Akt-p70 S6 kinase signaling pathway (Lin and Leonard, 2000; Malek and Castro, 2010), which promotes cell growth and survival (Franke et al., 1997). This pathway is inhibited by rapamycin, which blocks the function of a serine and threonine kinase, mTOR (mechanistic target of rapamycin), which is activated by PI-3K-AKT pathway and (Thomson et al., 2009). STAT5A and STAT5B dock on IL-2Rβ, are phosphorylated, dimerize, and translocate to the nucleus, where they bind to target genes essential for effector cell function and T cell growth (Friedmann et al., 1996; Lin et al., 2012) as well as differentiation (Liao et al., 2011; Liao et al., 2008). Generally, IL-2 is viewed as an activator, inducing more genes than it represses (Lin et al., 2012). Interestingly, a microRNA, miR-182, that is induced by IL-2 is an inhibitor of the transcription factor FOXO1, which blocks cell cycle progression in resting T cells, thereby helping to mediate the clonal expansion of activated helper T cells (Stittrich et al., 2010).
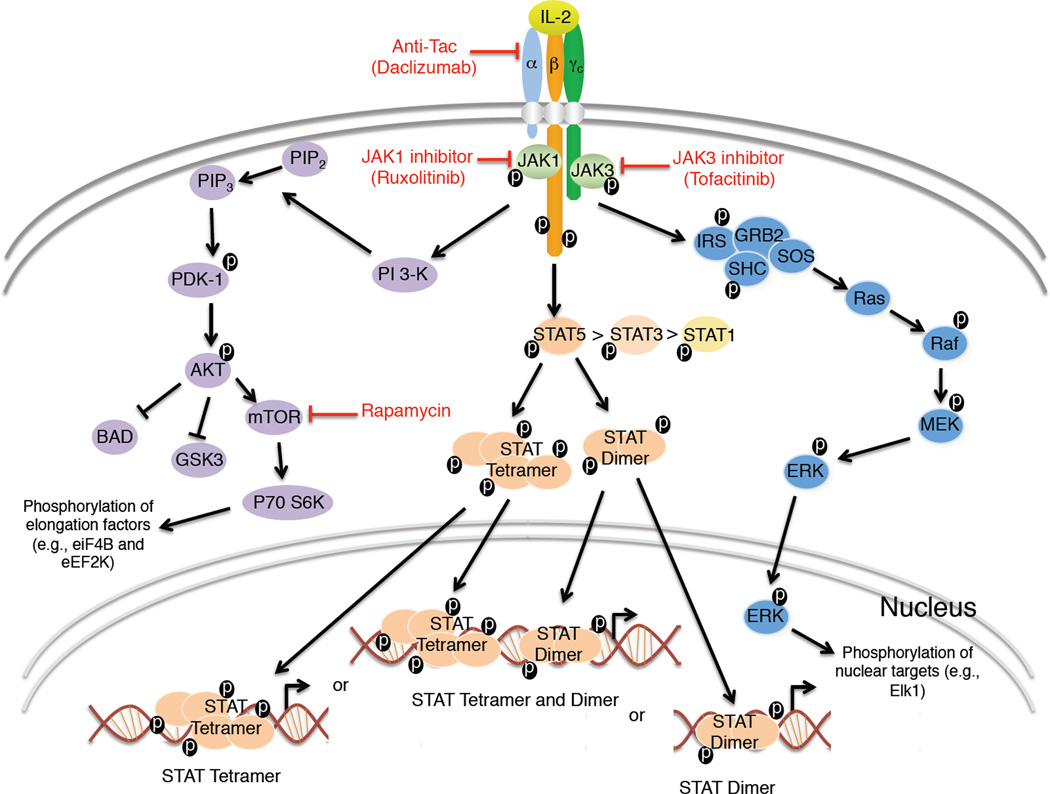
Figure 4. Schematic of major IL-2 signaling pathways.
Shown is the activation of PI 3-K-AKT, JAK-STAT, and SHC-RAS-MAPK signaling pathways. Also shown are potential therapeutic points of control for IL-2 signaling, with anti-Tac (daclizumab), rapamycin, and JAK1 or JAK3 inhibitors being shown in red. The cartoon shows signaling by both STATs dimers and tetramers. The figure indicates that IL-2 activates more STAT5 than STAT3 and more STAT3 than STAT1. ERK refers to both ERK1 and ERK2. MEK refers to both MEK1 and MEK2.
In addition to their tyrosine phosphorylation-mediated dimerization, like STAT1 and STAT4 (Chen et al., 2003), STAT5 proteins can interact via their N-terminal region (N-domains), allowing tetramer or higher order oligomer formation and binding to tandem GAS motifs (Lin et al., 2012). The importance of STAT5 tetramers has been analyzed by generating mice in which Il28 and Phe81 in the STAT5 N-domain have been mutated to alanines in the Stat5a, Stat5b, or both the Stat5a and Stat5b genes (Lin et al., 2012), allowing formation of STAT5 dimers but not tetramers. ChIP-Seq and gene expression analyses of cells from wild-type and mutant mice has allowed the identification of genes regulated by STAT5 dimers versus tetramers and the definition of dimer versus tetramer consensus motifs (Lin et al., 2012). Defective expression of certain genes, such as Il2ra, in the tetramerization-defective mice, can readily be attributed to known STAT5 tetramer binding sites, and these mice have decreased NK cell and CD8+ T cells and defective gene expression in response to IL-2 or IL-15. Moreover, these animals exhibit diminished virus-specific CD8+ T cell expansion, particularly early after infection with lymphocytic choriomeningitis virus (LCMV) or adenovirus 5, indicating that STAT5 tetramers are more important for early responses than for the later memory development and maintenance (Lin et al., 2012). An adoptive-transfer colitis model reveals that normal Treg cell-mediated suppression is also dependent on STAT5 tetramers (Lin et al., 2012), consistent with their requirement for IL-2 signaling (Malek and Castro, 2010; Malek et al., 2002). Thus, IL-2 function requires STAT5 tetramers as well as dimers.
IL2RG, X-linked severe combined immunodeficiency, and a major conundrum
Studies of the IL-2 receptor γ chain has revealed that the gene encoding this receptor protein, IL2RG, is located at the disease locus for X-linked severe combined immunodeficiency (XSCID) at Xq13 (Noguchi et al., 1993b). XSCID is a profound inherited immunodeficiency affecting approximately 1 in every 200,000 live births in the United States, representing the most common inherited immunodeficiency. XSCID is characterized by essentially absent T and NK cells; B cell numbers are relatively normal but are non-functional. Without successful bone marrow transplantation, affected individuals typically die in the first year of life (Cavazzana-Calvo et al., 2005; Leonard, 1996). Strikingly, IL2RG (encoding IL-2Rγ) is mutated in humans with XSCID (Noguchi et al., 1993b). This finding was most unexpected, given normal T and NK cell development in humans with IL-2 deficiency or in Il2−/− mice, leading to the hypothesis that IL-2Rγ has roles beyond IL-2 (Noguchi et al., 1993b). Indeed, IL-2Rγ was initially shown to be a critical component for the IL-4 and IL-7 receptors and subsequently for IL-9, IL-15, and IL-21 as well, hence its being renamed as the common cytokine receptor γ chain, γc (Leonard, 2001; Noguchi et al., 1993a)(Figure 1). The knowledge that JAK3 physically associates with γc led to speculation and confirmation that mutations in JAK3 caused a similar clinical and immunological disease to XSCID (Macchi et al., 1995; Russell et al., 1995). The identification of IL7R-deficient SCID patients with a selective T cell defect (Puel et al., 1998) indicates that the T cell defect in XSCID results from defective IL-7 signaling; the ability of IL-15 to drive NK development (Waldmann, 2006) indicates that defective IL-15 signaling explains the lack of NK cells (Leonard, 1996); and analysis of mice in which both the Il21r and Il4 genes were deleted revealed that defective signaling by the combination of IL-4 and IL-21 explained the defective B cell function in XSCID (Ozaki et al., 2002). Thus, although studies of IL-2 led to the discovery of the cause of XSCID, defective IL-2 signaling is not responsible for the major defects in that disease.
Consistent with the conclusion that defective IL-15 signaling is responsible for the lack of NK cells in XSCID and JAK3 deficiency, defective expression of IL-2Rβ, which is shared by the receptors for IL-2 and IL-15, results in an NK-deficient form of SCID (Gilmour et al., 2001)(see Figure 1). Interestingly, mutation of IL2RA results not in SCID but an autoimmune syndrome (Sharfe et al., 1997) (see Figure 1), consistent with a selective defect in IL-2 signaling, which is required for normal Treg cell development (Malek and Castro, 2010). As expected, like humans with IL2RB deficiency, Il2rb−/− mice also lack NK cells, and like humans with IL2RA deficiency, Il2ra−/− mice exhibit an autoimmune syndrome. Interestingly, Il2rg−/− mice have a major defect not only in T and NK cells but also in B cells, given the requirement for IL-7 for B cell development in mice but not humans (Leonard, 2001). These animals do not exhibit autoimmunity given the profound developmental defects in T, B, and NK cells (Leonard, 2001).
IL-2 and IL-15: two closely related but biologically distinct cytokines
As discussed above, IL-2 and IL-15 are highly related to each other and both cytokines bind to receptors that contain IL-2Rβ and γc to transduce their signals, although they have distinct α chains (Waldmann, 2006). Their three dimensional quaternary structures reveal that IL-15 binds to the IL-2Rβ and γc heterodimer in nearly identical fashion to that of IL-2, with similar gene induction profiles (Ring et al., 2012), but yet they are very distinctive biologically (Waldmann, 2006) (Figure 1), based in part on distinctive patterns of expression for IL-2 and IL-15 as well as IL-2Rα vs. IL-15Rα. Despite the fact that both IL-2 and IL-15 share IL-2Rβ and γc as receptor components and activate common signaling pathways, they exhibit signaling differences (Castro et al., 2011; Cornish et al., 2006). For example, in freshly isolated CD8+ splenic T cells, IL-15 is more potent than IL-2 in driving cell cycle progression and more effectively cooperates with IL-21 than does IL-2 (Zeng et al., 2005), whereas in antigen-activated CD8+ T cells IL-2 more potently increases protein synthesis and cell proliferation than does IL-15 (Cornish et al., 2006). This effect of IL-2 correlates with rapid expansion of short-lived effector cells, whereas IL-15 favors the maintenance of long-lived, memory phenotype cells (Waldmann, 2006). As noted above, both cytokines can use trans-presentation although this mechanism is more utilized by IL-15.
Because of the sharing of IL-2Rβ, a monoclonal antibody (Mikβ1) to this protein can inhibit not only IL-2 signaling but also IL-15 signaling. Indeed, it was reported that abnormal IL-15 responses might contribute to the survival and expansion of certain leukemias and/or lymphomas and moreover, Mikβ1 blocked trans-presentation of IL-15 and now is being used in a phase I clinical trial for T-cell large granular lymphocytic leukemia (T-LGL) (Waldmann et al., 2012).
IL-2 and Treg cells
Regulatory T (Treg) cells play vital roles in preventing autoimmunity, limiting inflammatory responses and maintaining immune homeostasis (Littman and Rudensky, 2010; Sakaguchi et al., 2008; Shevach, 2009). A hallmark of Treg cells is expression of the forkhead transcription factor FOXP3, which is necessary but not sufficient for Treg development (Figure 5). TCR-induced CpG hypomethylation of conserved noncoding DNA sequences (CNSs) critically regulates FOXP3 expression and is important for establishing Treg cell identity (Kim and Leonard, 2007; Ohkura et al., 2012; Zheng et al., 2010). Most FOXP3+ Treg cells are CD4+CD25+ cells (Sakaguchi et al., 2008; Shevach, 2009). In addition to thymic-derived natural Tregs cells, naïve peripheral CD4+ T cells can become CD4+FOXP3+ inducible Treg cells when stimulated with anti-CD3, TGF-β, and IL-2, with a critical role for IL-2 in their generation and expansion (Davidson et al., 2007; Zheng et al., 2007). The suppressive actions of Treg cells in part can be attributed to their ability to efficiently compete with effector CD4+ T cells for the available IL-2 (Busse et al., 2010; Pandiyan et al., 2007). Interestingly, this can result in apoptosis of the effector cells, in part through an increased expression of pro-apoptotic protein BIM and a decreased phosphorylation of the pro-apoptotic protein BAD due to lower AKT activity (Pandiyan et al., 2007).
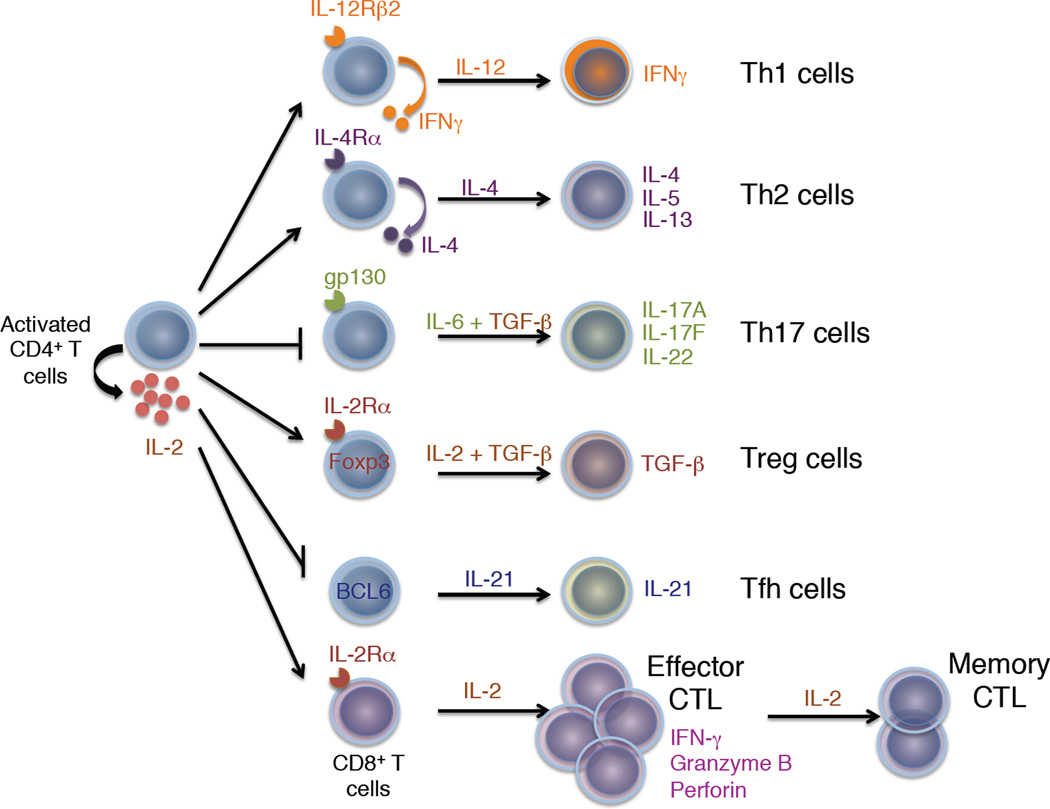
Figure 5. IL-2 is important in many types of T effector cell differentiation.
Shown is the induction by IL-2 of IL-12Rβ2 to promote Th1 cell differentiation, of IL-4Rα to promote Th2 cell differentiation, and of IL-2Rα to promote Treg cell differentiation. Conversely, IL-2 represses expression of gp130 (and IL-6Rα) while inducing T-bet (not shown) to repress Th17 cell differentiation. IL-2 is also a repressor of Tfh differentiation based on its repression of BCL6 expression. Finally, IL-2 promotes the differentiation, expansion, and the cytolytic activity of cytotoxic T cells.
An essential role of IL-2 in controlling immune homeostasis was first suggested by the spontaneous lethal autoimmunity found in mice lacking expression of IL-2, IL-2Rα, or IL-2Rβ (Leonard, 2001). Adoptive transfer of wild type CD4+CD25+ T cells into Il2rb−/− mice prevents autoimmunity, demonstrating a critical role for IL-2 for Treg cell development and function in vivo (Antony et al., 2006; Malek and Castro, 2010; Malek et al., 2002). Consistent with this, in the non-obese diabetic (NOD) mouse model of type 1 diabetes, intra-islet Treg cell dysfunction due to defective IL-2 production results in the breakdown of self-tolerance, but administering low doses of IL-2 and IL-2 mAbs (Boyman et al., 2006), which form stable immune complexes, increased Treg cell survival and decreased the incidence of diabetes (Tang et al., 2008), without activating effector CD8+ T cells. Similarly, pretreatment of mice with IL-2-anti-IL-2 complexes boosts Treg cell numbers and ameliorates experimental allergic encephalitis, myasthenia gravis, and graft rejection (Boyman and Sprent, 2012). Beneficial effects of IL-2 has also observed in approximately half of patients with glucocorticoid refractory chronic graft-versus-host disease, with expansion of Treg but not conventional T cells (Koreth et al., 2011). Similarly, low-dose IL-2 increased Treg cells in patients with autoimmune vasculitis secondary to hepatitis C virus infection (Saadoun et al., 2011).
IL-2 and T helper cell differentiation
CD4+ T cells can differentiate into multiple helper T cell populations upon antigen exposure, including Th1, Th2, Th9, Th17, and Tfh cells (Crotty, 2011; Jabeen and Kaplan, 2012; Littman and Rudensky, 2010; Paul and Zhu, 2010; Szabo et al., 2003) (Figure 5). IL-12 drives the differentiation of Th1 cells, which produce IFN-γ; IL-4 drives differentiation of Th2 cells, which produce IL-4, IL-5, and IL-13; the combination of IL-4 and TGFβ drives differentiation of Th9 cells which produce IL-9; IL-6 + TGF-β drives differentiation of Th17 cells, which produce IL-17A, IL-17F, and IL-22; and IL-6 + IL-21 promotes the differentiation of Tfh cells. These differentiation pathways are not necessarily terminal, however, and subsets of Th cells can acquire the ability to produce other Th cell-specific cytokine(s) (O'Shea and Paul, 2010).
Th1 cell differentiation
Th1 cells mediate host defense to viruses and intracellular pathogens and contribute to the development of pathogenic inflammatory diseases based on their production of IFN-γ. IL-12 via STAT4 is essential for driving Th1 cell differentiation and inducing T-bet, promoting the survival and proliferation of Th1 cells, a transcriptionally permissive chromatin structure at the Ifng locus, and enhanced IL-12Rβ2 expression (Afkarian et al., 2002; Szabo et al., 2003; Zhang and Boothby, 2006). IL-2 induces IFN-γ, both by inducing T cell proliferation (Bird et al., 1998) and via a STAT5-dependent mechanism (Shi et al., 2008). Moreover, IL-2 via STAT5 also rapidly induces IL-12Rβ2 (Liao et al., 2011), augmenting responsiveness to IL-12. Retroviral expression of IL-12Rβ2 could restore diminished Th1 cell differentiation in Il2−/− T cells; thus, IL-2-induced IL-12Rβ2 is critical for Th1 cell differentiation. IL-2 also induces Tbx21, which encodes T-bet, but T-bet alone could not restore Th1 cell differentiation (Liao et al., 2011). An essential role for IL-2 in Th1 cell differentiation in vivo has also been reported using Il27r−/− mice. These mice have elevated production of IL-2, and when infected with T. gondii, they developed a lethal inflammatory disease associated with increased IFN-γ that is diminished by IL-2 blockade (Villarino et al., 2006).
Th2 cell differentiation
Th2 cells control humoral immunity to extracellular parasites and mediate allergic inflammatory responses (Paul and Zhu, 2010). In vitro Th2 differentiation requires the TCR-mediated activation of naïve CD4+ T cells in the presence of anti-IFN-γ, IL-2, and IL-4, with IL-2 promoting chromatin accessibility at the Il4 locus (Cote-Sierra et al., 2004). TCR stimulation induces IL-4Rα expression by an IL-2 and STAT5-dependent mechanism, promoting cellular responsiveness to IL-4, and transduction of Il4ra into Il2-deficient T cells can rescue Th2 cell differentiation in the absence of IL-2 (Liao et al., 2008). IL-2-induced STAT5 binds throughout the Il4-Il13-Il5 Th2 cell-associated cytokine locus cluster, at hypersensitive site 2 (HS2), HS3, and HS5, as well as within the locus control region B and C elements in the Rad50 gene (Liao et al., 2008). Thus, IL-2 via STAT5A and STAT5B directly regulates IL-4Rα and IL-4 expression to promote Th2 cell differentiation. Interestingly, other cytokines that activate STAT5, including IL-7 and IL-15, also induce IL-4Rα on T cells, suggesting that depending on the cellular context, other cytokines can also prime CD4+ T cells for Th2 cell differentiation (Liao et al., 2008).
Th17 cell differentiation
Th17 cells produce IL-17A, IL-17F, and IL-22, which are important for anti-bacterial and anti-fungal immunity, as well as in autoimmune diseases, including multiple sclerosis, psoriasis, diabetes, rheumatoid arthritis, and Crohn’s disease (Littman and Rudensky, 2010). Differentiation of Th17 cells can be induced by IL-6 + TGF-β via an IL-6 to IL-23 to IL-21 cytokine cascade (Littman and Rudensky, 2010). IL-2 signaling decreases Th17 cell generation, and when immunized, host mice receiving adoptively transferred Il2−/− CD4+ T cells generate more Th17 cells than did mice receiving wild type cells (Laurence et al., 2007). It has been proposed that IL-2-activated STAT5 competes with STAT3 for binding to sites in the Il17a locus, thus inhibiting Il17a transcription (Yang et al., 2011), but direct STAT5 inhibition of Il17a transcription has not been shown. IL-2 also inhibits expression of both IL-6Rα and gp130, with increased expression of these receptors in Il2−/− T cells (Liao et al., 2011), and retroviral transduction of Il6st, which encodes gp130, partially overcomes IL-2-induced inhibition of IL-17A production (Liao et al., 2011). Nevertheless, IL-2 partially inhibits IL-17A even when gp130 is constitutively expressed, indicating that IL-2 also inhibits Th17 cell differentiation by a receptor-independent mechanism (Liao et al., 2011). In this regard, IL-2 also induces expression of Tbx21, which inhibits Th17 cell differentiation (Liao et al., 2011), consistent with the ability of T-bet to interact with Runx1 and to prevent association of Runx1 with RORγt, which is essential for Th17 differentiation (Lazarevic et al., 2011). Interestingly, in Th17 cells, Tbx21 retroviral transduction increases IFN-γ production, suggesting that IL-2 stimulation can promote a Th17-to-Th1 cell shift in these cells (Liao et al., 2011). Although IL-2 can inhibit Th17 cell differentiation, adults recently infected with HIV-1 maintain peripheral Th17 cell numbers when treated with IL-2 (Ndhlovu et al., 2010), and IL-2 can expand Th17 cells once generated, which helps to explain benefits of anti-IL-2 receptor based immunotherapy in uveitis and scleritis, where Th17 cells are pathogenic (Amadi-Obi et al., 2007). Thus, IL-2 has complex actions on Th17 cells, inhibiting their differentiation but promoting their expansion.
Tfh cell differentiation
T follicular helper (Tfh) cells reside in germinal centers and promote immunoglobulin class switch and antibody affinity maturation by B cells (Crotty, 2011). Tfh cells are CD4+ T cells and both secrete and respond to IL-21, augmenting their expression of ICOS, CXCR5 and BCL-6. IL-2 via STAT5 induces BLIMP1, which suppresses Tfh differentiation and germinal center formation (Ballesteros-Tato et al., 2012; Johnston et al., 2012); IL-2 also inhibits BCL6 in Th1 cells, which suppresses a Tfh-like gene profile (Oestreich et al., 2012).
IL-2 and effector-memory cytolytic CD8+ T cell differentiation
Besides its actions on Th cell populations, IL-2 drives development of naïve CD8+ T cells into effector and memory cytolytic T-lymphocytes (CTL) upon antigen stimulation in the context of infection or inflammation, with induction of IFN-γ, perforin, and granzymes (Pipkin et al., 2010). IL-2 increases expression of the transcription factor Eomes, which together with STAT5 upregulates Prf1 expression (Pipkin et al., 2010). IL-2 also potently suppresses BCL6 and IL-7Rα (Pipkin et al., 2010; Xue et al., 2002). Resting CD8+ T cells express IL-2Rβ and γc, so concentrations of IL-2 sufficient to titrate intermediate affinity receptors can induce proliferation of these cells, which interestingly requires formation of STAT5 tetramers (Lin et al., 2012), as well as the proliferation of activated CD8+ T cells that express high affinity IL-2 receptors.
In LCMV infection, in contrast to WT cells, Il2ra−/− CD8+ T cells do not expand to secondary antigen exposure even though they robustly respond to primary viral infection, indicating a critical role for IL-2 in development of the memory response (Williams et al., 2006). During infection, CD25 expression is initially high but then becomes heterogeneous; cells that downregulate CD25 early in infection receive a less prolonged or “intense” IL-2 signal, upregulate IL-7Rα and CD62L, and eventually become long-lived memory cells, whereas cells expressing higher numbers of CD25 molecules undergo more rapid proliferation, exhibit a terminally differentiated effector phenotype, and are eliminated by apoptosis (Kalia et al., 2010; Williams et al., 2006). A critical role for endogenous IL-2 production in the antiviral response has also been demonstrated using Il2−/− mice (Cousens et al., 1995). Moreover, autocrine production of IL-2 by CD8+ T cells is important for the development of memory CD8+ T cells in vivo (Feau et al., 2011). Overall, these studies support the model that both the duration and strength of the IL-2 signal is important, with contributions of IL-2 to the generation of effector cells as well as memory cell differentiation.
IL-2 and Adult T-Cell Leukemia-Lymphoma (ATL)
Human T cell lymphotropic virus, type 1 (HTLV-1) is a retrovirus that causes adult T cell leukemia-lymphoma (ATL) and tropical spastic paraparesis-HTLV-1 associated myelopathy (TSP-HAM). In the early phase of ATL, there is a period of autocrine growth of leukemic CD4+ T cells, with expression of IL-2 and functional IL-2 receptors, and anti-Tac mAb to human IL-2Rα can decrease proliferation of these cells during this phase of ATL (Waldmann et al., 2001). Over time, however, IL-2 production is lost, although cell surface IL-2Rα expression persists, and this later phase is associated with IL-2-independent growth. Interestingly, ATL cells exhibit Treg cell-like suppressor activity (Waldmann et al., 1984). In many patients, this later phase is associated with a constitutively-activated JAK-STAT pathway (Migone et al., 1995). Humanized anti-Tac (daclizumab or Zenopax)(Figure 4), was approved by the FDA to prevent renal allograft rejection but has also been used to treat patients with ATL (Waldmann, 2007) and relapsing-remitting multiple sclerosis (Martin, 2012); whereas the antibody alone was effective in a very limited number of individuals, coupling the antibody to immunotoxin-linked mAbs or arming it with 90Y are approaches that have been used to enhance effectiveness (Waldmann, 2007).
IL-2 as a therapeutic modality
IL-2 immunotherapy has been used for many years, with FDA approval in 1992 for metastatic renal cell carcinoma and in 1998 for metastatic melanoma. IL-2 treatment can result in complete remission of 5–10% of patients with these diseases, with lack of recurrence for as long as 25 years and potential cures of 70% of these individuals, with complete tumor regression (Rosenberg, 2012). However, this represents only a small fraction of the patients. Vaccination with a melanoma peptide vaccine can significantly improve effectiveness (Schwartzentruber et al., 2011), and the addition of adoptive cell transfer approaches have improved cure rates for metastatic melanoma to between 20 and 40% (Rosenberg, 2012). Such treatment involves the isolation of tumor infiltrating lymphocytes (TILs) grown from a patient’s tumor and expansion of these autologous TILs prior to their reintroduction (Rosenberg, 2012). A limitation of IL-2 is its toxicity, including severe capillary leak syndrome that can accompany such treatment. In mice, endothelial nitric oxide synthase appears to play a critical role in the development of IL-2-induced capillary leak syndrome (Samlowski et al., 2011).
Interestingly, although IL-2 is a potent T-cell growth factor, in animal models in which adoptive immunotherapy has been performed, IL-2 is less effective as an anti-tumor agent than IL-21, which inhibits expression of IL-2Rα on CD8+ T cells, potentially decreasing the responsiveness of cells to IL-2 (Hinrichs et al., 2008). The decreased anti-tumor effect has been potentially attributed to the more potent induction of terminal effector CTL differentiation by IL-2, whereas IL-21-treated CD8+ T cells persist for longer periods of time in vivo.
Recently, “super-IL-2” variants of IL-2 have been developed that exhibit enhanced binding to IL-2Rβ and do not require IL-2Rα (Levin et al., 2012). As compared to wild type IL-2, they induce greater expansion of cytotoxic T cells but less expansion of Treg cells, with reduced pulmonary edema in mice when administered in vivo (Levin et al., 2012). Based on the crystal structure, the mutations that result in the augmented function are mainly in the core structure and stabilize IL-2, reducing the flexibility of a helix in the IL-2Rβ binding site, and resulting in a conformation that is normally achieved only in the presence of IL-2Rα (Levin et al., 2012). Presumably for this reason, the IL-2 superkine induces vigorous signaling and phosphorylation in the absence IL-2Rα (Levin et al., 2012). More studies are needed to determine if this or another engineered IL-2 has therapeutic advantages over wild type IL-2.
Inhibition of IL-2 signaling as a therapeutic modality
In addition to dicluzamib, other modes of IL-2 inhibition are possible. As noted above, IL-2 activates JAK1 and JAK3, with JAK1 associating with IL-2Rβ and JAK3 with γc (Russell et al., 1994), and it was proposed that inhibitors of JAK3 would be immunosuppressive (Russell et al., 1995). Two JAK inhibitors are now available for clinical use. Ruxolitinib (Jakafi) inhibits JAK1 > JAK2 (Harrison et al., 2012) and has been approved by the FDA for treating myelofibrosis, with ongoing clinical trials for primary polycythemia and primary thrombocythemia. Tofacitinib (Changelian et al., 2003; Kremer et al., 2009), which inhibits JAK3 > JAK1 and JAK2 and has also been approved for the treatment of rheumatoid arthritis, based on successful clinical trials (Kontzias et al., 2012), with ongoing Phase III clinical trials for the treatment of psoriasis (Figure 4). Other JAK inhibitors are being evaluated for use in rheumatoid arthritis and other diseases as well. It is important to note that these agents inhibit not only IL-2, but also other cytokines that utilized the inhibited JAK kinases. Finally, IL-2 mutants with inhibitory activity as well as structural based medicinal chemistry approaches (Wilson and Arkin, 2011) may also potentially yield useful inhibitors of IL-2.
Conclusions and perspective
As discussed herein, IL-2 mediates diverse pleiotropic actions, promoting T-cell proliferation, survival, cytolytic activity, NK cell activity, development of Treg cells, and AICD. Moreover, by inducing IL-12Rβ2 and IL-4Rα but suppressing gp130 expression, IL-2 increases responsiveness to IL-12 and IL-4 to promote Th1 and Th2 cell differentiation while inhibiting the IL-6 to IL-23 to IL-21 cytokine cascade that drives Th17 cell differentiation, and furthermore, its induction of IL-2Rβ not only increases responsiveness to IL-2 but also to IL-15, with potential impact on CTL and NK activity. Thus, IL-2 does not by itself determine the eventual differentiation but rather facilitates or inhibits responsiveness to other cytokines, in part by promoting or inhibiting different cytokine cascades. Consistent with its potent proliferative activity for T cells, IL-2 promotes the expansion of multiple populations, including even Th17 cells whose differentiation it inhibits. The amount of IL-2 produced, when and where IL-2 receptors are expressed, the affinities (high- or intermediate-affinity) of these receptors, and the other cytokines present in the cellular milieu, are critical in determining the biological outcome orchestrated by IL-2 during an immune response. Moreover, different populations of responding cells can potentially compete for IL-2 and Treg cells expressing high affinity IL-2Rs can efficiently compete for limited amount IL-2 with effector cells that express intermediate affinity IL-2Rs (Busse et al., 2010; Pandiyan et al., 2007). Furthermore, when IL-2 production by effector cells is abrogated during acute massive inflammation in lethal T. gondii infection, there is a marked decrease in Treg cell numbers and function that was reversed by exogenous IL-2 (Oldenhove et al., 2009). Another complexity is the possible competition among γc family cytokines. For example, if one γc family cytokine is more abundant than others, it might quantitatively recruit γc to its receptor, thereby effectively sequestering γc and diminishing cellular responsiveness to the other cytokines. Overall, we have tremendous knowledge regarding how IL-2 and its receptor are regulated as well as related to its structure, its signaling pathways, and its relationship to the five other cytokines that share γc. These basic research findings have led to the development of new therapeutic approaches potentially applicable to a range of diseases, either based on the administration of IL-2 or by using antagonists of IL-2 production or signaling. How and when it is desirable to modulate the IL-2 response is context-dependent. For example, promoting effector activity may be a desired response for treatment of cancer, whereas blocking effector responses or augmenting Treg cell activity may be beneficial in preventing autoimmunity or massive inflammatory responses, as noted above related to T. gondii. Thus, directed manipulation of the activities of this cytokine, which has major actions both related to effector T cells and in tolerance, could have implications for controlling complex immune responses and for immunotherapy of a range of diseases. Moreover, future studies directed towards comprehensively mapping the IL-2 signaling network and further elucidating the molecular mechanisms of IL-2 action could facilitate the development of better agonists and more specific antagonists to efficiently modulate immune responses.
Acknowledgments
We thank Drs. Rosanne Spolski, Chi-Keung Wan, and Erin B. West, all in NHLBI, for critical comments. This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Amadi-Obi A, Yu CR, Liu X, Mahdi RM, Clarke GL, Nussenblatt RB, Gery I, Lee YS, Egwuagu CE. TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat Med. 2007;13:711–718. doi: 10.1038/nm1585. [DOI] [PubMed] [Google Scholar]
- Antony PA, Paulos CM, Ahmadzadeh M, Akpinarli A, Palmer DC, Sato N, Kaiser A, Hinrichs CS, Klebanoff CA, Tagaya Y, Restifo NP. Interleukin-2-dependent mechanisms of tolerance and immunity in vivo. J Immunol. 2006;176:5255–5266. doi: 10.4049/jimmunol.176.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima N, Kamio M, Imada K, Hori T, Hattori T, Tsudo M, Okuma M, Uchiyama T. Pseudo-high affinity interleukin 2 (IL-2) receptor lacks the third component that is essential for functional IL-2 binding and signaling. J Exp Med. 1992;176:1265–1272. doi: 10.1084/jem.176.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci U S A. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- Bosco MC, Espinoza-Delgado I, Schwabe M, Gusella GL, Longo DL, Sugamura K, Varesio L. Regulation by interleukin-2 (IL-2) and interferon gamma of IL-2 receptor gamma chain gene expression in human monocytes. Blood. 1994;83:2995–3002. [PubMed] [Google Scholar]
- Boussiotis VA, Barber DL, Nakarai T, Freeman GJ, Gribben JG, Bernstein GM, D'Andrea AD, Ritz J, Nadler LM. Prevention of T cell anergy by signaling through the gamma c chain of the IL-2 receptor. Science. 1994;266:1039–1042. doi: 10.1126/science.7973657. [DOI] [PubMed] [Google Scholar]
- Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- Busse D, de la Rosa M, Hobiger K, Thurley K, Flossdorf M, Scheffold A, Hofer T. Competing feedback loops shape IL-2 signaling between helper and regulatory T lymphocytes in cellular microenvironments. Proc Natl Acad Sci U S A. 2010;107:3058–3063. doi: 10.1073/pnas.0812851107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Kozak CA, Liu YJ, Noguchi M, O'Connell E, Leonard WJ. Characterization of cDNAs encoding the murine interleukin 2 receptor (IL-2R) gamma chain: chromosomal mapping and tissue specificity of IL-2R gamma chain expression. Proc Natl Acad Sci U S A. 1993;90:8464–8468. doi: 10.1073/pnas.90.18.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro I, Yu A, Dee MJ, Malek TR. The basis of distinctive IL-2- and IL-15-dependent signaling: weak CD122-dependent signaling favors CD8+ T central-memory cell survival but not T effector-memory cell development. J Immunol. 2011;187:5170–5182. doi: 10.4049/jimmunol.1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Lagresle C, Hacein-Bey-Abina S, Fischer A. Gene therapy for severe combined immunodeficiency. Annu Rev Med. 2005;56:585–602. doi: 10.1146/annurev.med.56.090203.104142. [DOI] [PubMed] [Google Scholar]
- Changelian PS, Flanagan ME, Ball DJ, Kent CR, Magnuson KS, Martin WH, Rizzuti BJ, Sawyer PS, Perry BD, Brissette WH, et al. Prevention of organ allograft rejection by a specific Janus kinase 3 inhibitor. Science. 2003;302:875–878. doi: 10.1126/science.1087061. [DOI] [PubMed] [Google Scholar]
- Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE, Jr, Kuriyan J. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 2003;12:361–365. doi: 10.1110/ps.0218903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikuma S, Suita N, Okazaki IM, Shibayama S, Honjo T. TRIM28 prevents autoinflammatory T cell development in vivo. Nat Immunol. 2012;13:596–603. doi: 10.1038/ni.2293. [DOI] [PubMed] [Google Scholar]
- Cornish GH, Sinclair LV, Cantrell DA. Differential regulation of T-cell growth by IL-2 and IL-15. Blood. 2006;108:600–608. doi: 10.1182/blood-2005-12-4827. [DOI] [PubMed] [Google Scholar]
- Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci U S A. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens LP, Orange JS, Biron CA. Endogenous IL-2 contributes to T cell expansion and IFN-gamma production during lymphocytic choriomeningitis virus infection. J Immunol. 1995;155:5690–5699. [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V, Macino G. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115:265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- Depper JM, Leonard WJ, Drogula C, Kronke M, Waldmann TA, Greene WC. Interleukin 2 (IL-2) augments transcription of the IL-2 receptor gene. Proc Natl Acad Sci U S A. 1985;82:4230–4234. doi: 10.1073/pnas.82.12.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depper JM, Leonard WJ, Kronke M, Noguchi PD, Cunningham RE, Waldmann TA, Greene WC. Regulation of interleukin 2 receptor expression: effects of phorbol diester, phospholipase C, and reexposure to lectin or antigen. J Immunol. 1984;133:3054–3061. [PubMed] [Google Scholar]
- Driesen J, Popov A, Schultze JL. CD25 as an immune regulatory molecule expressed on myeloid dendritic cells. Immunobiology. 2008;213:849–858. doi: 10.1016/j.imbio.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Feau S, Arens R, Togher S, Schoenberger SP. Autocrine IL-2 is required for secondary population expansion of CD8(+) memory T cells. Nat Immunol. 2011;12:908–913. doi: 10.1038/ni.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- Friedmann MC, Migone TS, Russell SM, Leonard WJ. Different interleukin 2 receptor beta-chain tyrosines couple to at least two signaling pathways and synergistically mediate interleukin 2-induced proliferation. Proc Natl Acad Sci U S A. 1996;93:2077–2082. doi: 10.1073/pnas.93.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour KC, Fujii H, Cranston T, Davies EG, Kinnon C, Gaspar HB. Defective expression of the interleukin-2/interleukin-15 receptor beta subunit leads to a natural killer cell-deficient form of severe combined immunodeficiency. Blood. 2001;98:877–879. doi: 10.1182/blood.v98.3.877. [DOI] [PubMed] [Google Scholar]
- Giri JG, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park LS, Cosman D, Anderson D. Utilization of the beta and gamma chains of the IL-2 receptor by the novel cytokine IL-15. Embo J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- Granucci F, Vizzardelli C, Pavelka N, Feau S, Persico M, Virzi E, Rescigno M, Moro G, Ricciardi-Castagnoli P. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat Immunol. 2001;2:882–888. doi: 10.1038/ni0901-882. [DOI] [PubMed] [Google Scholar]
- Gruss HJ, Scott C, Rollins BJ, Brach MA, Herrmann F. Human fibroblasts express functional IL-2 receptors formed by the IL-2R alpha- and beta-chain subunits: association of IL-2 binding with secretion of the monocyte chemoattractant protein-1. J Immunol. 1996;157:851–857. [PubMed] [Google Scholar]
- Harrison C, Verstovsek S, McMullin MF, Mesa R. Janus kinase inhibition and its effect upon the therapeutic landscape for myelofibrosis: from palliation to cure? Br J Haematol. 2012;157:426–437. doi: 10.1111/j.1365-2141.2012.09108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemar A, Subtil A, Lieb M, Morelon E, Hellio R, Dautry-Varsat A. Endocytosis of interleukin 2 receptors in human T lymphocytes: distinct intracellular localization and fate of the receptor alpha, beta, and gamma chains. J Cell Biol. 1995;129:55–64. doi: 10.1083/jcb.129.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko AY, Suzuki R, Charles N, Alvarez-Errico D, Sargent JL, Laurence A, Rivera J. Mast cell interleukin-2 production contributes to suppression of chronic allergic dermatitis. Immunity. 2011;35:562–571. doi: 10.1016/j.immuni.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs CS, Spolski R, Paulos CM, Gattinoni L, Kerstann KW, Palmer DC, Klebanoff CA, Rosenberg SA, Leonard WJ, Restifo NP. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood. 2008;111:5326–5333. doi: 10.1182/blood-2007-09-113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabeen R, Kaplan MH. The symphony of the ninth: the development and function of Th9 cells. Curr Opin Immunol. 2012;24:303–307. doi: 10.1016/j.coi.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–366. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Kim HP, Kim BG, Letterio J, Leonard WJ. Smad-dependent cooperative regulation of interleukin 2 receptor alpha chain gene expression by T cell receptor and transforming growth factor-beta. J Biol Chem. 2005;280:34042–34047. doi: 10.1074/jbc.M505833200. [DOI] [PubMed] [Google Scholar]
- Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontzias A, Kotlyar A, Laurence A, Changelian P, O'Shea JJ. Jakinibs: a new class of kinase inhibitors in cancer and autoimmune disease. Curr Opin Pharmacol. 2012;12:464–470. doi: 10.1016/j.coph.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koreth J, Matsuoka K, Kim HT, McDonough SM, Bindra B, Alyea EP, 3rd, Armand P, Cutler C, Ho VT, Treister NS, et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. N Engl J Med. 2011;365:2055–2066. doi: 10.1056/NEJMoa1108188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JM, Bloom BJ, Breedveld FC, Coombs JH, Fletcher MP, Gruben D, Krishnaswami S, Burgos-Vargas R, Wilkinson B, Zerbini CA, Zwillich SH. The safety and efficacy of a JAK inhibitor in patients with active rheumatoid arthritis: Results of a double-blind, placebo-controlled phase IIa trial of three dosage levels of CP-690,550 versus placebo. Arthritis Rheum. 2009;60:1895–1905. doi: 10.1002/art.24567. [DOI] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M, Chan KM, Hornung F, McFarland H, Siegel R, Wang J, Zheng L. Mature T lymphocyte apoptosis--immune regulation in a dynamic and unpredictable antigenic environment. Annu Rev Immunol. 1999;17:221–253. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- Leonard WJ. The molecular basis of X-linked severe combined immunodeficiency: defective cytokine receptor signaling. Annu Rev Med. 1996;47:229–239. doi: 10.1146/annurev.med.47.1.229. [DOI] [PubMed] [Google Scholar]
- Leonard WJ. Cytokines and immunodeficiency diseases. Nat Rev Immunol. 2001;1:200–208. doi: 10.1038/35105066. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Depper JM, Crabtree GR, Rudikoff S, Pumphrey J, Robb RJ, Kronke M, Svetlik PB, Peffer NJ, Waldmann TA, et al. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature. 1984;311:626–631. doi: 10.1038/311626a0. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Depper JM, Uchiyama T, Smith KA, Waldmann TA, Greene WC. A monoclonal antibody that appears to recognize the receptor for human T-cell growth factor; partial characterization of the receptor. Nature. 1982;300:267–269. doi: 10.1038/300267a0. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Kronke M, Peffer NJ, Depper JM, Greene WC. Interleukin 2 receptor gene expression in normal human T lymphocytes. Proc Natl Acad Sci U S A. 1985;82:6281–6285. doi: 10.1073/pnas.82.18.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ, Shores EW, Love PE. Role of the common cytokine receptor gamma chain in cytokine signaling and lymphoid development. Immunol Rev. 1995;148:97–114. doi: 10.1111/j.1600-065x.1995.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, Su L, Moraga I, Raeber ME, Bowman GR, Novick P, et al. Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'. Nature. 2012;484:529–533. doi: 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JX, Bhat NK, John S, Queale WS, Leonard WJ. Characterization of the human interleukin-2 receptor beta-chain gene promoter: regulation of promoter activity by ets gene products. Mol Cell Biol. 1993;13:6201–6210. doi: 10.1128/mcb.13.10.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JX, Leonard WJ. The immediate-early gene product Egr-1 regulates the human interleukin-2 receptor beta-chain promoter through noncanonical Egr and Sp1 binding sites. Mol Cell Biol. 1997;17:3714–3722. doi: 10.1128/mcb.17.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur Rasheed M, Rochman Y, Wang L, Cui K, Liu C, et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 2012;36:586–599. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Lowenthal JW, Greene WC. Contrasting interleukin 2 binding properties of the alpha (p55) and beta (p70) protein subunits of the human high-affinity interleukin 2 receptor. J Exp Med. 1987;166:1156–1161. doi: 10.1084/jem.166.4.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O'Shea JJ, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- Martin R. Anti-CD25 (daclizumab) monoclonal antibody therapy in relapsing-remitting multiple sclerosis. Clin Immunol. 2012;142:9–14. doi: 10.1016/j.clim.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator, Fos, attenuating T cell proliferation and survival. J Exp Med. 2008;205:1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migone TS, Lin JX, Cereseto A, Mulloy JC, O'Shea JJ, Franchini G, Leonard WJ. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- Mingari MC, Gerosa F, Carra G, Accolla RS, Moretta A, Zubler RH, Waldmann TA, Moretta L. Human interleukin-2 promotes proliferation of activated B cells via surface receptors similar to those of activated T cells. Nature. 1984;312:641–643. doi: 10.1038/312641a0. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu ZJ, Oishi I, Silvennoinen O, Witthuhn BA, Ihle JN, et al. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- Morgan DA, Ruscetti FW, Gallo R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science. 1976;193:1007–1008. doi: 10.1126/science.181845. [DOI] [PubMed] [Google Scholar]
- Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Liu XW, Wynshaw-Boris A, Rosenthal LA, Imada K, Finbloom DS, Hennighausen L, Leonard WJ. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Russell SM, Mess SA, Friedmann M, Erdos M, Francois C, Jacques Y, Adelstein S, Leonard WJ. Heterodimerization of the IL-2 receptor beta- and gamma-chain cytoplasmic domains is required for signalling. Nature. 1994;369:330–333. doi: 10.1038/369330a0. [DOI] [PubMed] [Google Scholar]
- Naranjo-Gomez M, Oliva H, Climent N, Fernandez MA, Ruiz-Riol M, Bofill M, Gatell JM, Gallart T, Pujol-Borrell R, Borras FE. Expression and function of the IL-2 receptor in activated human plasmacytoid dendritic cells. Eur J Immunol. 2007;37:1764–1772. doi: 10.1002/eji.200636980. [DOI] [PubMed] [Google Scholar]
- Ndhlovu LC, Sinclair E, Epling L, Tan QX, Ho T, Jha AR, Eccles-James I, Tincati C, Levy JA, Nixon DF, et al. IL-2 immunotherapy to recently HIV-1 infected adults maintains the numbers of IL-17 expressing CD4+ T (T(H)17) cells in the periphery. J Clin Immunol. 2010;30:681–692. doi: 10.1007/s10875-010-9432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BH, Lord JD, Greenberg PD. Cytoplasmic domains of the interleukin-2 receptor beta and gamma chains mediate the signal for T-cell proliferation. Nature. 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- Nikaido T, Shimizu A, Ishida N, Sabe H, Teshigawara K, Maeda M, Uchiyama T, Yodoi J, Honjo T. Molecular cloning of cDNA encoding human interleukin-2 receptor. Nature. 1984;311:631–635. doi: 10.1038/311631a0. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, Leonard WJ. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993a;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993b;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, et al. T cell receptor stimulation-induced epigenetic changes and foxp3 expression are independent and complementary events required for treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, O'Brien S, Blank R, Lamb E, Natarajan S, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC, 3rd, Liu C, Schwartzberg PL, Leonard WJ. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298:1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- Paliard X, de Waal Malefijt R, Yssel H, Blanchard D, Chretien I, Abrams J, de Vries J, Spits H. Simultaneous production of IL-2, IL-4, and IFN-gamma by activated human CD4+ and CD8+ T cell clones. J Immunol. 1988;141:849–855. [PubMed] [Google Scholar]
- Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Peng SL, Gerth AJ, Ranger AM, Glimcher LH. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity. 2001;14:13–20. doi: 10.1016/s1074-7613(01)00085-1. [DOI] [PubMed] [Google Scholar]
- Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Ziegler SF, Buckley RH, Leonard WJ. Defective IL7R expression in T(−)B(+)NK(+) severe combined immunodeficiency. Nat Genet. 1998;20:394–397. doi: 10.1038/3877. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, Rangachari M, Zhu C, Xiao S, Seavitt J, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring AM, Lin JX, Feng D, Mitra S, Rickert M, Bowman GR, Pande VS, Li P, Moraga I, Spolski R, et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat Immunol. 2012;13:1187–1195. doi: 10.1038/ni.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb RJ, Munck A, Smith KA. T cell growth factor receptors. Quantitation, specificity, and biological relevance. J Exp Med. 1981;154:1455–1474. doi: 10.1084/jem.154.5.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003634. 127ps128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin LA, Nelson DL. The soluble interleukin-2 receptor: biology, function, and clinical application. Ann Intern Med. 1990;113:619–627. doi: 10.7326/0003-4819-113-8-619. [DOI] [PubMed] [Google Scholar]
- Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- Russell SM, Keegan AD, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann MC, Miyajima A, Puri RK, Paul WE, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-4 receptor. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- Saadoun D, Rosenzwajg M, Joly F, Six A, Carrat F, Thibault V, Sene D, Cacoub P, Klatzmann D. Regulatory T-cell responses to low-dose interleukin-2 in HCV-induced vasculitis. N Engl J Med. 2011;365:2067–2077. doi: 10.1056/NEJMoa1105143. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Samlowski WE, Kondapaneni M, Tharkar S, McGregor JR, Laubach VE, Salvemini D. Endothelial nitric oxide synthase is a key mediator of interleukin-2-induced hypotension and vascular leak syndrome. J Immunother. 2011;34:419–427. doi: 10.1097/CJI.0b013e31821dcb50. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kuhn R, Muller W, Palm N, Rude E. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- Schuh K, Twardzik T, Kneitz B, Heyer J, Schimpl A, Serfling E. The interleukin 2 receptor alpha chain/CD25 promoter is a target for nuclear factor of activated T cells. J Exp Med. 1998;188:1369–1373. doi: 10.1084/jem.188.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber DJ, Lawson DH, Richards JM, Conry RM, Miller DM, Treisman J, Gailani F, Riley L, Conlon K, Pockaj B, et al. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med. 2011;364:2119–2127. doi: 10.1056/NEJMoa1012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharfe N, Shahar M, Roifman CM. An interleukin-2 receptor gamma chain mutation with normal thymus morphology. J Clin Invest. 1997;100:3036–3043. doi: 10.1172/JCI119858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon M, Klausner RD, Cullen BR, Chizzonite R, Leonard WJ. Novel interleukin-2 receptor subunit detected by cross-linking under high-affinity conditions. Science. 1986;234:859–863. doi: 10.1126/science.3095922. [DOI] [PubMed] [Google Scholar]
- Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin TH, Appell KC, Berg LJ. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity. 2008;28:763–773. doi: 10.1016/j.immuni.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JP, Sharon M, Smith PL, Leonard WJ. The IL-2 receptor beta chain (p70): role in mediating signals for LAK, NK, and proliferative activities. Science. 1987;238:75–78. doi: 10.1126/science.3116668. [DOI] [PubMed] [Google Scholar]
- Stauber DJ, Debler EW, Horton PA, Smith KA, Wilson IA. Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proc Natl Acad Sci U S A. 2006;103:2788–2793. doi: 10.1073/pnas.0511161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- Takeshita T, Asao H, Ohtani K, Ishii N, Kumaki S, Tanaka N, Munakata H, Nakamura M, Sugamura K. Cloning of the gamma chain of the human IL-2 receptor. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E, Piccirillo CA, Salomon BL, Bluestone JA. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Matsui H, Fujita T, Takaoka C, Kashima N, Yoshimoto R, Hamuro J. Structure and expression of a cloned cDNA for human interleukin-2. Nature. 1983;302:305–310. doi: 10.1038/302305a0. [DOI] [PubMed] [Google Scholar]
- Teshigawara K, Wang HM, Kato K, Smith KA. Interleukin 2 high-affinity receptor expression requires two distinct binding proteins. J Exp Med. 1987;165:223–238. doi: 10.1084/jem.165.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele S, Wittmann J, Jack H-M, Pahl A. miR-9 enhances IL-2 production in activated human CD4+ T cells by repressing Blimp-1. Eur. J. Immunol. 2012;42:2100–2108. doi: 10.1002/eji.201142203. [DOI] [PubMed] [Google Scholar]
- Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudo M, Kozak RW, Goldman CK, Waldmann TA. Demonstration of a non-Tac peptide that binds interleukin 2: a potential participant in a multichain interleukin 2 receptor complex. Proc Natl Acad Sci U S A. 1986;83:9694–9698. doi: 10.1073/pnas.83.24.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Broder S, Waldmann TA. A monoclonal antibody (anti-Tac) reactive with activated and functionally mature human T cells. I. Production of anti-Tac monoclonal antibody and distribution of Tac (+) cells. J Immunol. 1981;126:1393–1397. [PubMed] [Google Scholar]
- Villarino AV, Stumhofer JS, Saris CJ, Kastelein RA, de Sauvage FJ, Hunter CA. IL-27 limits IL-2 production during Th1 differentiation. J Immunol. 2006;176:237–247. doi: 10.4049/jimmunol.176.1.237. [DOI] [PubMed] [Google Scholar]
- von Bergwelt-Baildon MS, Popov A, Saric T, Chemnitz J, Classen S, Stoffel MS, Fiore F, Roth U, Beyer M, Debey S, et al. CD25 and indoleamine 2,3-dioxygenase are up-regulated by prostaglandin E2 and expressed by tumor-associated dendritic cells in vivo: additional mechanisms of T-cell inhibition. Blood. 2006;108:228–237. doi: 10.1182/blood-2005-08-3507. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. Daclizumab (anti-Tac, Zenapax) in the treatment of leukemia/lymphoma. Oncogene. 2007;26:3699–3703. doi: 10.1038/sj.onc.1210368. [DOI] [PubMed] [Google Scholar]
- Waldmann TA, Conlon KC, Stewart DM, Worthy TA, Janik JE, Fleisher TA, Albert PS, Figg WD, Spencer SD, Raffeld M, et al. Phase I trial of IL-15 transpresentation blockade using humanized Mik-Beta-1 monoclonal antibody in patients with T-cell large granular lymphocytic leukemia. Blood. 2012 doi: 10.1182/blood-2012-08-450585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann TA, Dubois S, Tagaya Y. Contrasting roles of IL-2 and IL-15 in the life and death of lymphocytes: implications for immunotherapy. Immunity. 2001;14:105–110. [PubMed] [Google Scholar]
- Waldmann TA, Greene WC, Sarin PS, Saxinger C, Blayney DW, Blattner WA, Goldman CK, Bongiovanni K, Sharrow S, Depper JM, et al. Functional and phenotypic comparison of human T cell leukemia/lymphoma virus positive adult T cell leukemia with human T cell leukemia/lymphoma virus negative Sezary leukemia, and their distinction using anti-Tac. Monoclonal antibody identifying the human receptor for T cell growth factor. J Clin Invest. 1984;73:1711–1718. doi: 10.1172/JCI111379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HM, Smith KA. The interleukin 2 receptor. Functional consequences of its bimolecular structure. J Exp Med. 1987;166:1055–1069. doi: 10.1084/jem.166.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- Weitzel RP, Lesniewski ML, Haviernik P, Kadereit S, Leahy P, Greco NJ, Laughlin MJ. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. 2009;113:6648–6657. doi: 10.1182/blood-2008-09-181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher FJ, 3rd, Kant JA. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- Wilson CG, Arkin MR. Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. Curr Top Microbiol Immunol. 2011;348:25–59. doi: 10.1007/82_2010_93. [DOI] [PubMed] [Google Scholar]
- Wong JC, Lee SB, Bell MD, Reynolds PA, Fiore E, Stamenkovic I, Truong V, Oliner JD, Gerald WL, Haber DA. Induction of the interleukin-2/15 receptor beta-chain by the EWS-WT1 translocation product. Oncogene. 2002;21:2009–2019. doi: 10.1038/sj.onc.1205262. [DOI] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Wuest SC, Edwan JH, Martin JF, Han S, Perry JS, Cartagena CM, Matsuura E, Maric D, Waldmann TA, Bielekova B. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat Med. 2011;17:604–609. doi: 10.1038/nm.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Kovanen PE, Pise-Masison CA, Berg M, Radovich MF, Brady JN, Leonard WJ. IL-2 negatively regulates IL-7 receptor alpha chain expression in activated T lymphocytes. Proc Natl Acad Sci U S A. 2002;99:13759–13764. doi: 10.1073/pnas.212214999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Sharp LL, Havran WL, Rothenberg EV. Preferential activation of an IL-2 regulatory sequence transgene in TCR gamma delta and NKT cells: subset-specific differences in IL-2 regulation. J Immunol. 2004;172:4691–4699. doi: 10.4049/jimmunol.172.8.4691. [DOI] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, Hinrichs CS, Pise-Masison CA, Radonovich MF, Brady JN, Restifo NP, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med. 2005;201:139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006;203:1493–1505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA. IL-2 is essential for TGF-beta to convert naive CD4+CD25- cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J Immunol. 2007;178:2018–2027. doi: 10.4049/jimmunol.178.4.2018. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]