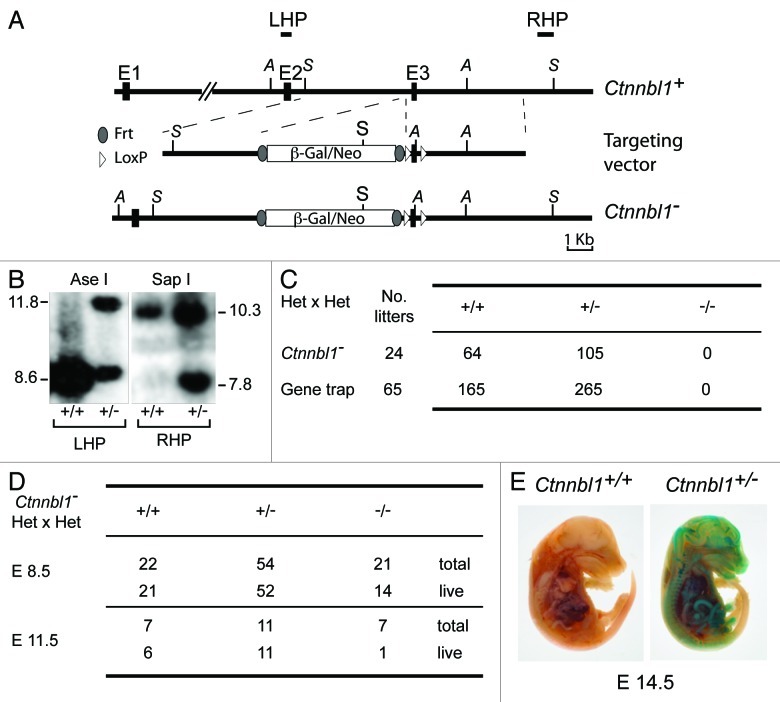
Figure 1. Targeted inactivation of Ctnnbl1 results in mid-term embryonic lethality. (A) Targeting the mouse Ctnnbl1 locus. The top line depicts the mouse Ctnnbl1 locus (three exons: E1, E2 and E3 are depicted) aligned with the targeting construct which contains a promoterless β-galactosidase/pgk-neo cassette flanked by flippase recognition sites (oval shapes) integrated into intron 2 with LoxP sites (open triangles) flanking E3. The left and right homology arms are 4.6 and 4.8 kb, respectively. Restriction endonuclease sites Ase I (A) and Sap I (S) are indicated as are the locations of the left- and right-hand probes (LHP and RHP) used for the Southern blot analysis. (B) Southern blot of tail DNA from mice carrying the targeted β-gal/neo cassette insertion into Ctnnbl1 E3 on one allele as well as controls hybridized with probes LHP and RHP. (C) Genotypes of weaned animals born to Ctnnnbl1+/− intercrosses. A similar analysis is shown for the progeny born to intercrosses of AX0016 mice that carry a gene trap insertion into the first Ctnnbl1 intron. (D) Genotypes of embryos obtained at day 8.5 or 11.5 of gestation from Ctnnnbl1+/− intercrosses. (E) Broad expression of the Ctnnbl1 locus as revealed by staining of a day 14.5 Ctnnnbl1+/− embryo for β-galactosidase activity.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.