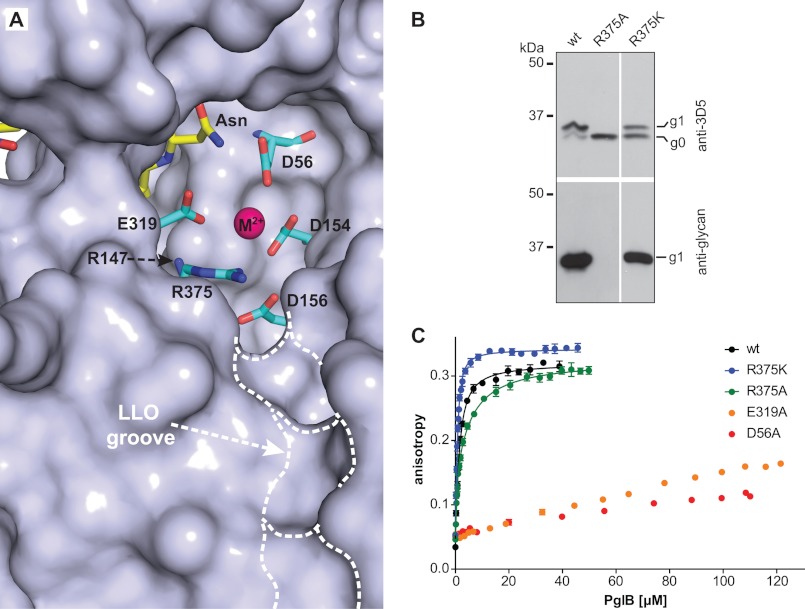
FIGURE 3.
Uncoupling peptide binding from glycosylation. A, surface representation of the active site of C. lari PglB (PDB code 3RCE). Catalytically important residues (cyan) and acceptor peptide (yellow) are shown in stick representation and are labeled. The bound divalent metal ion (M2+) is shown as a purple sphere. A hydrophobic groove possibly accommodating the isoprenoid part of LLO is indicated. B, immunoblots of in vivo glycosylation reactions detecting acceptor protein 3D5 (top) or bacterial N-glycans (bottom). Glycosylation results in a mobility shift from the non-glycosylated (g0) to the glycosylated form of the acceptor protein (g1). PglB mutants are indicated above the lanes. The lanes originate from the same gel and exposure time. C, fluorescence anisotropy experiments using fluorescently labeled peptide containing the DQNAT sequon, performed as described in the legend to Fig. 2B. The WT data are identical to that shown in Fig. 2B. Curve fitting was performed assuming a single binding site for all curves (R2(wt) = 0.9940; R2(R375K) = 0.9934; R2(R375A) = 0.9881). For the R375A mutant, a two-site model yielded a slightly better fit (R2 = 0.9951).