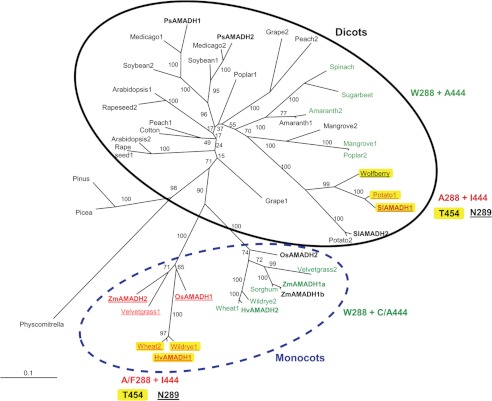
FIGURE 8.
Phylogenetic analysis of the plant ALDH10 family (AMADHs). The unrooted phylogenetic consensus tree shows two distinct groups of AMADHs, one from dicots (solid line) and the other from monocots (dashed line). PsAMADH numbering is used to point out crucial residues modulating substrate specificity. AMADHs exhibiting a significant BADH activity are highlighted in green. AMADHs predictable as broad substrate specific enzymes are highlighted in yellow due to the presence of Thr-454. The presence of Asn-289 implies that Tyr-163 is mobile, affecting the substrate binding. The enzymes from maize (ZmAMADHs) and tomato (SlAMADHs) analyzed in this work are shown in boldface type, as are the previously characterized reference enzymes from peas (PsAMADHs) (4), rice (OsAMADHs) (6), and barley (HvAMADHs) (18). The internal labels give bootstrap frequencies for each clade. See “Experimental Procedures” for the corresponding GenBankTM accession numbers.