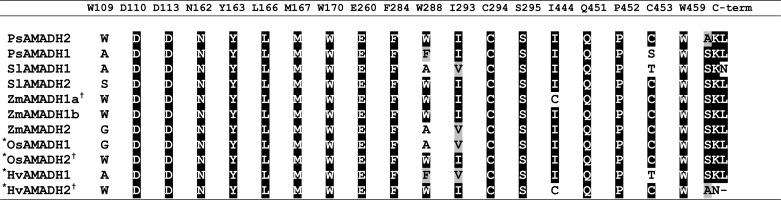
TABLE 5.
A comparison of active-site residues in ALDH10 family from several plant species
Shown are active-site residues and C-terminal motifs of AMADHs from pea (PsAMADH) (4), tomato (SlAMADH), and maize (ZmAMADH). The numbering used follows the sequence of PsAMADH2, of which the substrate channel was previously analyzed by site-directed mutagenesis (17). Rice (OsAMADHs, UniProtKB numbers O24174 and Q84LK3) and barley enzymes (HvAMADHs, UniProtKB numbers Q94IC0 and Q94IC1) analyzed by other groups (6, 18) are shown for comparison and indicated by an asterisk. Enzymes accepting BAL as an efficient substrate are labeled with a cross.