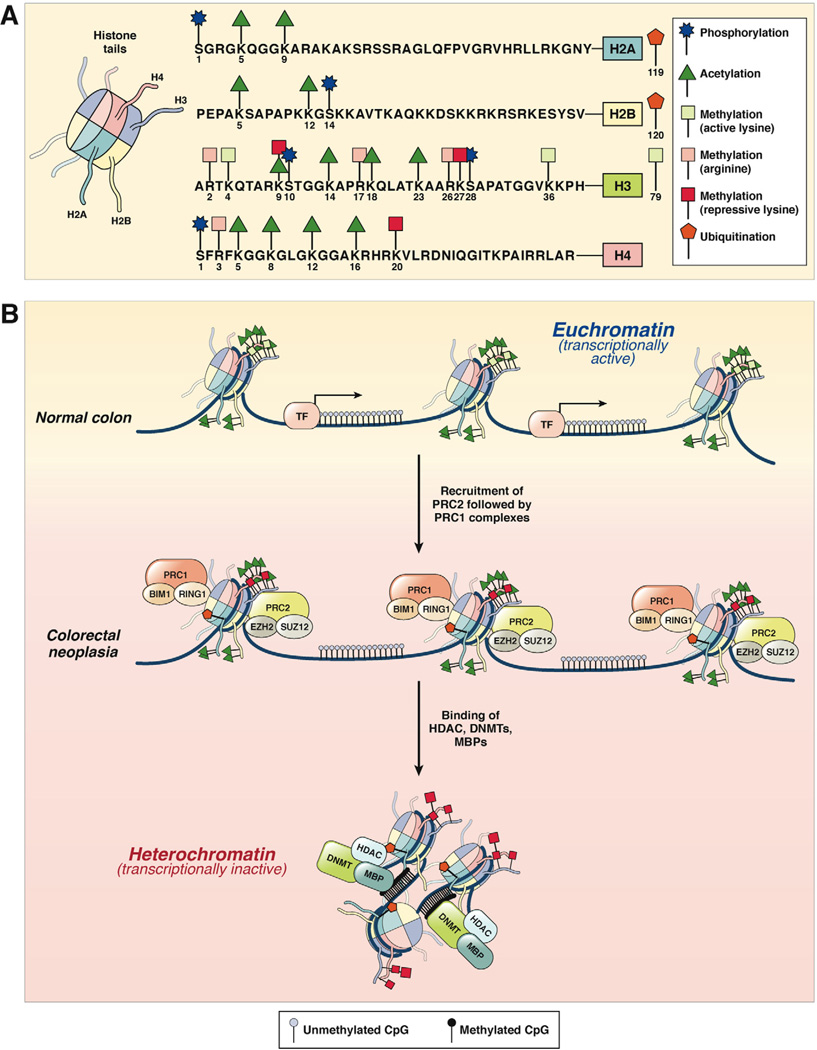
Figure 2.
(A) Histone modification patterns in colorectal neoplasia. The 4 histone proteins (H2A, H2B, H3, and H4), each with 2 copies, associate in cylindrical structures that constitute the histone core, and each nucleosome is a chromatin unit composed of 150 to 200 base pairs of DNA tightly wrapped around the octameric histone core. Covalent modifications of each histone tail, including acetylation, methylation, phosphorylation, and ubiquitination, are also shown on the right. The amino acid sequences of the N-terminal tails and the key selected sites of modification of these 4 histones are shown. (B) In normal colon, histones are arranged in the open, euchromatin configuration, which represents transcriptionally active chromatin (upper panel). Histone tails may undergo multiple modifications, which include histone acetylation (green triangles) and lysine methylation (green squares). Gene promoters in euchromatin allow easy access for transcription factor (TF) binding, permitting active gene expression. In CRC cells, chromatin configuration is converted to the more compacted heterochromatin configuration, which is transcriptionally inactive and results in the hypermethylation-induced silencing of genes. This process is facilitated by the multimeric polycomb repressive protein complexes, PRC2 and PRC1, which sequentially bind to histones and initiate histone methylation (initiated by PRC2 complex containing EZH2 and SUZ12) followed by maintenance methylation and monoubiquitination (performed by PRC1 complex with BIM1 and RING1 proteins). This is followed by the recruitment of DNMTs, HDACs, and methyl binding proteins (MBPs) to complete the PRC-mediated transcriptional silencing of genes.