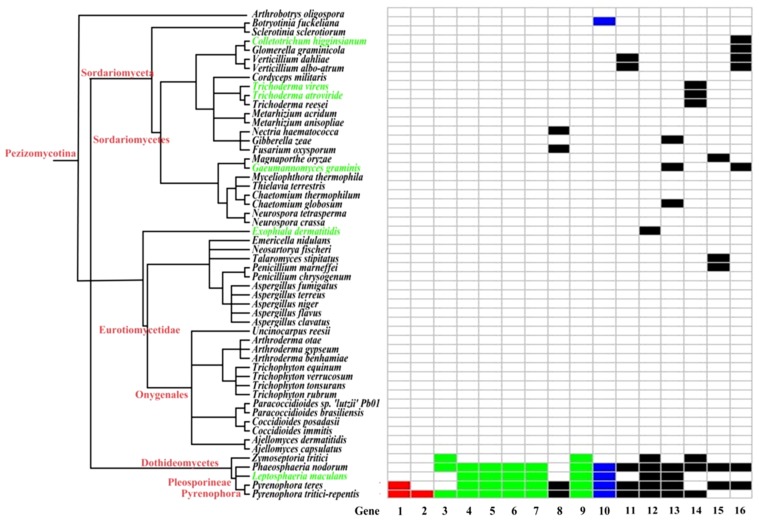
Figure 3. The presence or absence of HGT genes in fungi.
Species tree constructed from the taxonomy database of NCBI along with superfamily membership. Species with HGT genes but without genomic data are indicated in green. Boxes indicate possession of the gene. Red boxes: Genes limited to Pyrenophora, indicating a recent gene transfer event after divergence of Pyrenophora. Green boxes: Genes limited to Dothideomycetes, indicating a relatively ancient gene transfer event after divergence of Dothideomycetes. Blue boxes: Genes form two fungus clades that nest within non-fungal species in the corresponding gene tree, indicating two independent interkingdom transfers. Black boxes: Genes were present in Pyrenophora and distantly related organisms. Gene trees indicated independent interkingdom transfers. The trees cannot determine the presence of fungi–fungi HGT. Genes 1–16 refer to the genes encoding leucine-rich repeat protein, methyltransferase MppJ, beta-galactosidase, UDP-glucosyltransferase, GCN5-related N-acetyltransferase, oxidoreductase, Gfo/Idh/MocA family, enterochelin esterase-like enzyme, N-acetylglucosaminyltransferase, succinylglutamate desuccinylase/aspartoacylase, 5-formyltetrahydrofolate cyclo-ligase, NmrA family protein, glcG protein, xylanase A, cyanophycinase, alpha/beta hydrolase, oxidoreductase, respectively.