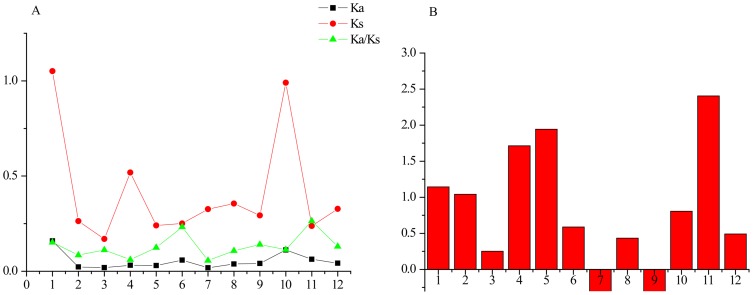
Figure 4. Selection pressure and evolutionary rates of HGTs in species of Pyrenophora.
(A) Nonsynonymous (Ka) and synonymous (Ks) mutations and their ratio Ka/Ks for the HGTs among species of Pyrenophora. (B) Comparison of molecular evolutionary rates between transferred and corresponding donor genes. Ka/Ks values of HGT genes in species of Pyrenophora were divided by Ka/Ks values of their corresponding homologs in donor group species, and the distribution of the log ratios was plotted. Values less than 0 indicated that transferred genes had lower Ka/Ks values than the corresponding donor genes. Values greater than 0 indicated that transferred genes had higher Ka/Ks values than corresponding donor genes. Genes 1–12 refer to genes encoding leucine-rich repeat protein, UDP-glucosyltransferase, GCN5-related N-acetyltransferase, oxidoreductase, Gfo/Idh/MocA family, enterochelin esterase-like enzyme, N-acetylglucosaminyltransferase, succinylglutamate desuccinylase/aspartoacylase, 5-formyltetrahydrofolate cyclo-ligase, NmrA family protein, glcG protein, xylanase A, cyanophycinase, respectively.