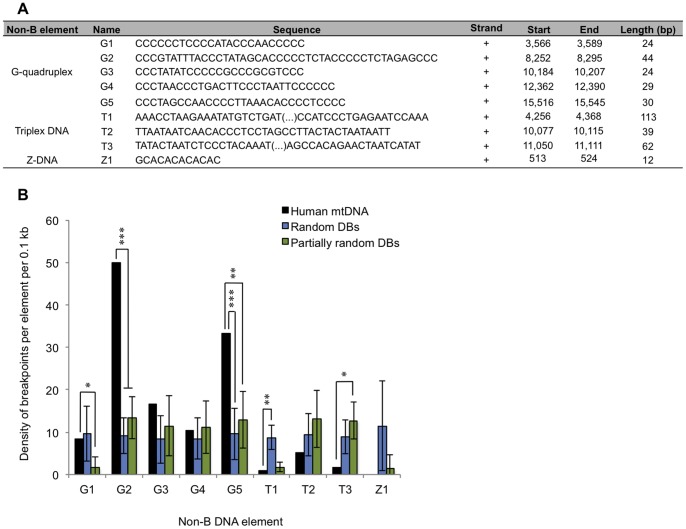
Figure 3. Impact of the presence of non-canonical (non-B) DNA structures on the distribution of deletion breakpoints.
(A) Sequences and locations of the DNA strings predicted to fold into G-quadruplexes, triplexes and Z-DNA. (B) Density of deletion breakpoints per 0.1 kb bins of each non-B element (black bars). Also shown are the density values computed after randomization of breakpoint positions (shown in blue) or after partially randomization of breakpoint positions (shown in green). Error bars represent standard deviations. * p<0.05; ** p<0.01; *** p<0.001.