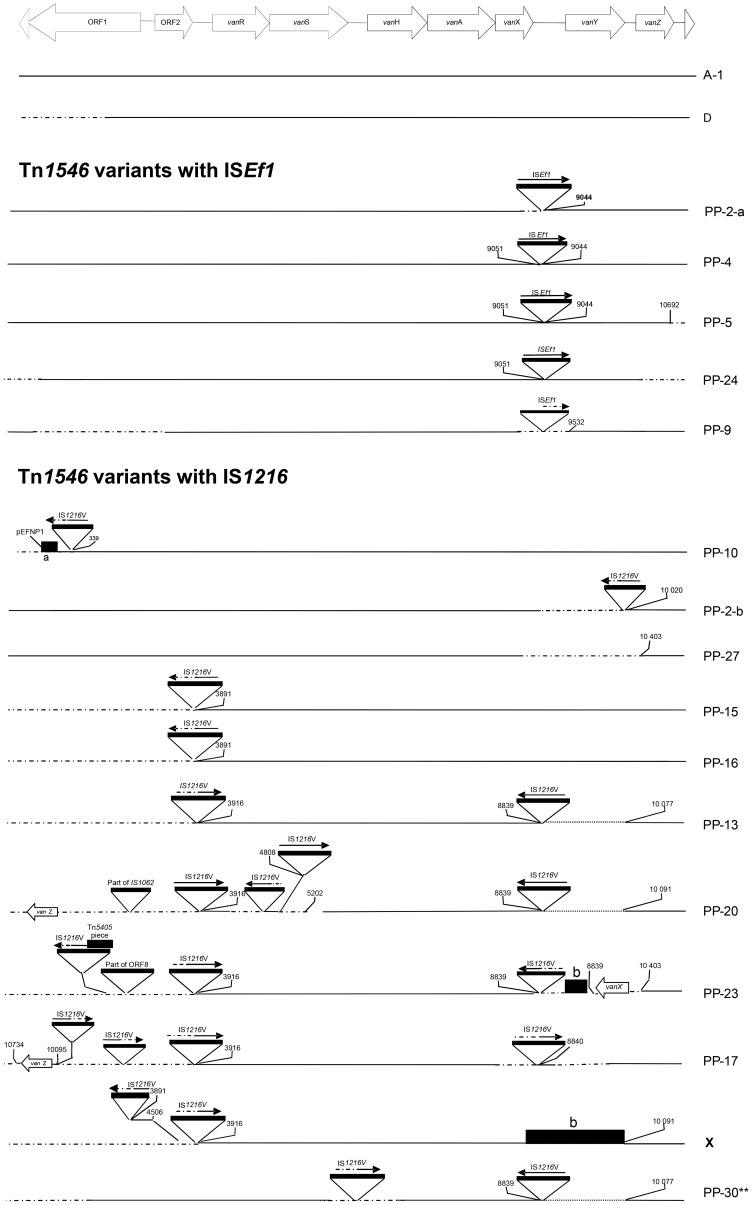
Figure 4. Genetic maps of Tn1546 variants.
Tn1546 variants are represented as previously described by Novais et al. [27] although grouped differently and specific types have been further explored (PP10, PP30): Tn1546 prototype A corresponds to the original sequence described by Arthur et al. [77] and D corresponds to Tn1546 variants from animals. Tn1546 variants with ISEf1 within vanX-vanY intergenic region (PP2a, PP4, PP5, PP9, PP24) and Tn1546 variants with IS1216 insertions at different positions (PP10, PP2b, PP13, PP15, PP16, PP17, PP20, PP23, PP27, PP30, X) are represented. The positions of genes and open reading frames and the direction of transcription are depicted with open arrows. IS elements are represented by triangles; other sequences are designated by rectangles. DNA insertions are represented highlighting the first nucleotide upstream and downstream from the insertion sites whenever known. Deletions are indicated by dots and discontinuous lines indicate sequences that were not characterized. (a) DNA sequence with homology to ORF3 (unknown protein product) and ORF1 (replication protein) of pEFNP1 plasmid (GenBank accession number AB038522). (b) DNA sequence with no match to any sequence available in GenBank. (*) PP23 was identified in an isolate susceptible to teicoplanin; this variant contained an insertion in the vanY gene that would affect the transcription of vanZ and it might explain the susceptibility to this glycopeptide as previously reported [27]. (**) PP30 was identified in an ST78 isolate susceptible to both glycopeptides (MIC against vancomycin and teicoplanin of 4 mg/L) carrying vanA-Tn1546. This variant contained alterations within the vanS-vanH intergenic region (an IS1216 insertion), which is involved in the expression and regulation of the resistance to vancomycin, and it constitutes the first description of a vanA isolate phenotypically susceptible to vancomycin in Portugal.