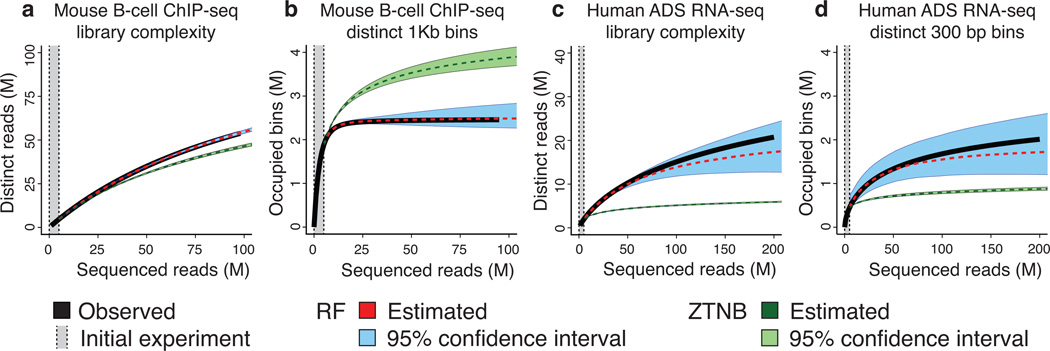
Figure 2.
Library complexity can be estimated both in terms of distinct molecules sequenced and in terms of distinct loci identified. (a) A ChIP-seq library (CTCF; mouse B-Cells) yields additional molecules after sequencing 100 million (M) reads; the RF remains accurate while the ZTNB loses accuracy. (b) In the same library, the number of mapped distinct genomic 1 kb windows saturates after 25 M reads. The rational function approximation (RF) is accurate and forecasts saturation, while the zero-truncated Negative Binomial (ZTNB) significantly overestimates. (c) An RNA-seq (Human adipose-derived mesenchymal stem (ADS) cells) library continues to yield additional molecules after 200 M reads; the RF remains accurate while the ZTNB predicts saturation. (d) In the same library, reads continued mapping to new 300 bp windows after 200 M reads. ZTNB incorrectly predicts saturation, while RF does not.