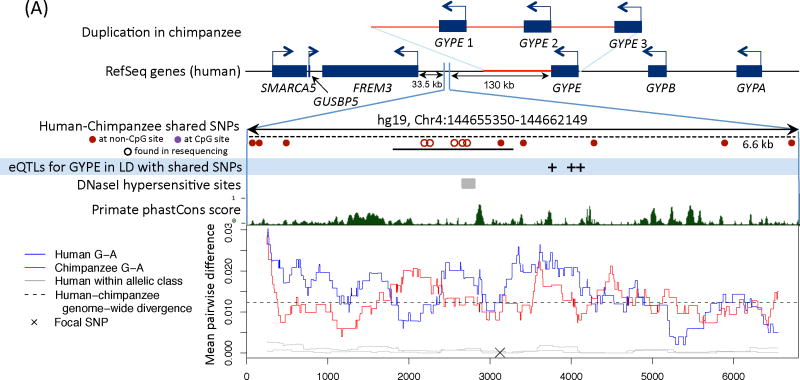
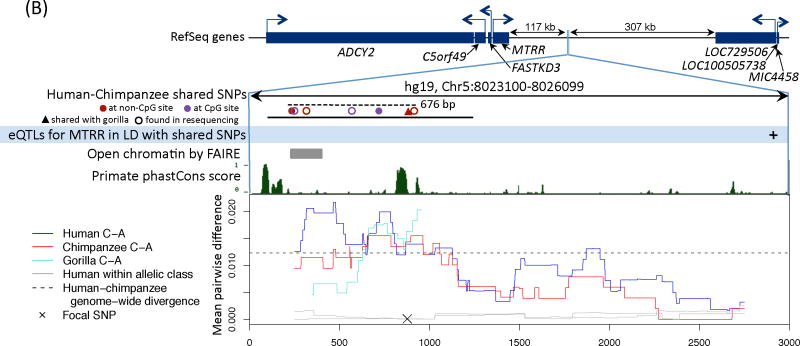
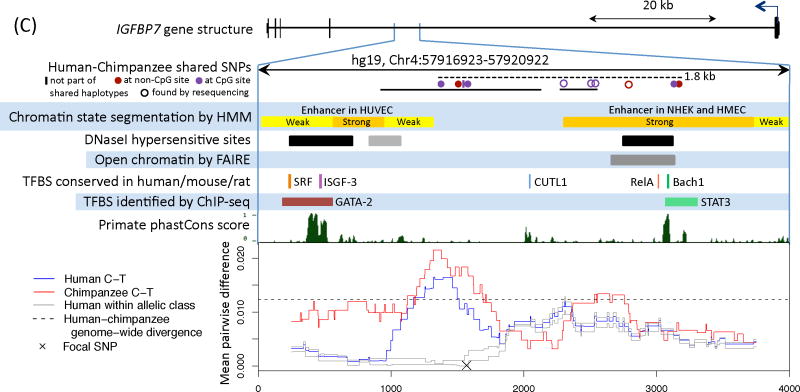
Figure 2. Functional information for three regions with a polymorphism shared identical by descent in humans and chimpanzees.
We show the nearby genes and direction of transcription, then a close up of the region with shared polymorphisms between humans and chimpanzees. The original shared SNPs used to identify shared haplotypes are shown as solid circles. The region resequenced in the validation experiment is indicated with a solid black bar and the length of the shared haplotypes with a dashed black bar (16). For sources of the functional annotation tracks shown, see (16). In the last panel, we focus on a shared SNP (hg19, chr4:144658471, chr5:8023976, and chr4:57918492, respectively) and show the average pairwise difference between allelic classes for humans (in blue) and chimpanzees (in red), for a 500 bp sliding window; the average pairwise difference within an allelic class in humans is in gray. We further indicate the average genome-wide divergence between human and chimpanzee (1.2%; (39)) with a dotted black line. For divergence between more distant ape species and a zoom out of diversity levels in each region, see Figs. S7 and S9.
A) FREM3. A duplication in chimpanzees that includes the GYPE gene is shown above the gene structure in humans (26). The shared SNPs and eQTLs for GYPE in monocytes (40) are in almost perfect LD, with a pairwise r2 ranging from 0.98 to 1.
B) MTRR. The shared SNP represented by a triangle is also seen in a sample of seven gorillas obtained by Sanger resequencing (see (16)); pairwise differences between allelic classes in gorillas is shown in turquoise for the resequenced region. The maximum pairwise r2 between a shared SNP and the eQTL for MTRR in monocytes is 0.47 (40) (16). The FAIRE signal is enriched in six cell lines.
C) IGFBP7. In the scan for shared haplotypes, five shared SNPs were found in the four kb region, occurring in two clusters with three and two SNPs, respectively, which are not in LD with each other in humans. Two of the shared SNPs found in the resequencing and a SNP outside the resequenced region constitute an additional instance of shared haplotypes. The FAIRE signal is enriched in four cell lines. Using a focal SNP in the second cluster yields similar results (see Fig. S7).