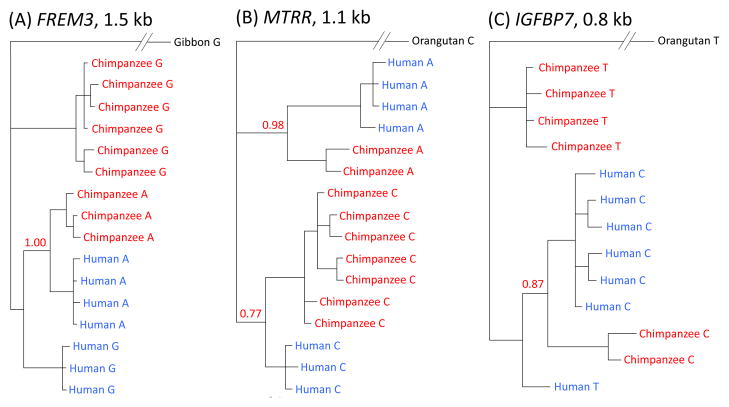
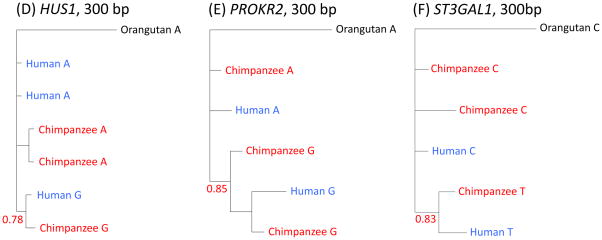
Figure 3.
Phylogenetic trees of haplotypes labeled with the same focal SNP considered in Fig. 2 or Fig. S8 for (A) FREM3, (B) MTRR, (C) IGFBP7, (D) HUS1, (E) PROKR2 and (F) ST3GAL1. Trees were generated from our resequencing data using MrBayes, with the median posterior probability of the clade over two runs reported in red (16). Results are for the entire resequenced regions for FREM3 and MTRR, and for the largest regions for which we found strong support in other cases. For FREM3, MTRR and IGFBP7, the regions on which the trees are based are long (>800 bps), providing strong support for a polymorphism shared identical by descent (16). For HUS1, the tree still clusters by allele when considering 1 kb (with posterior probability 0.58), but for ST3GAL1 and PROKR2, this is not the case (for more details, see (16)).