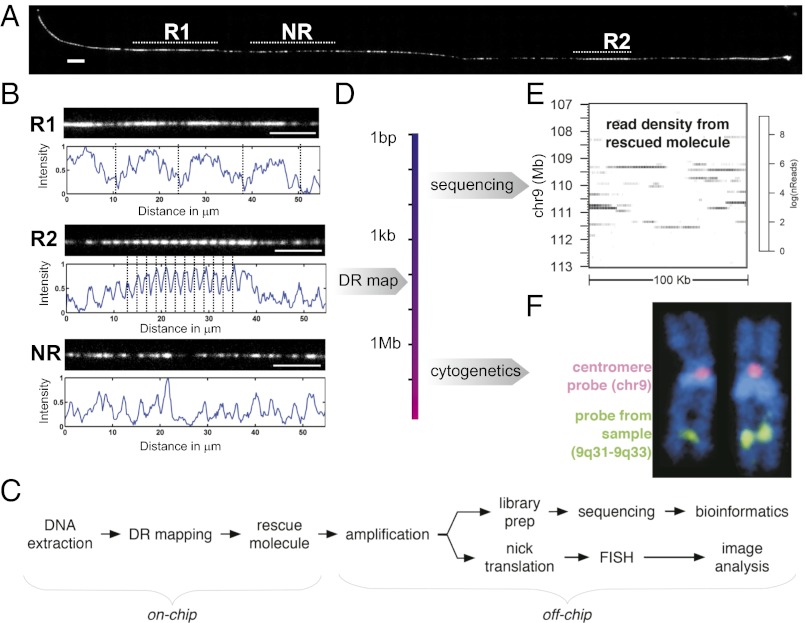
Fig. 4.
Workflow of integrated DR mapping and conventional genomics. (A) DR map of a single molecule from the Jurkat genome with two repeat features (R1, R2). A nonrepeat (NR) element is shown for comparison. Unique barcodes from the region between R1 and R2 failed to find significant match to any chromosomes in hg18, but reveals (B) that long-range repetitive elements are present. (Scale bar: 10 μm.) (C) A molecule is rescued and amplified, and sequencing and FISH analysis provides complementary information (D). (E) Sequencing reads are mapped to the reference genome to identify the origin of the rescued molecule as well as SNPs and short insertions/deletions (SI Appendix), and (F) FISH provides a bird’s-eye chromosomal view and confirmation of the origin identified by sequencing.