Figure 5. MICALs are Multi-domain Cytosolic Redox Enzymes.
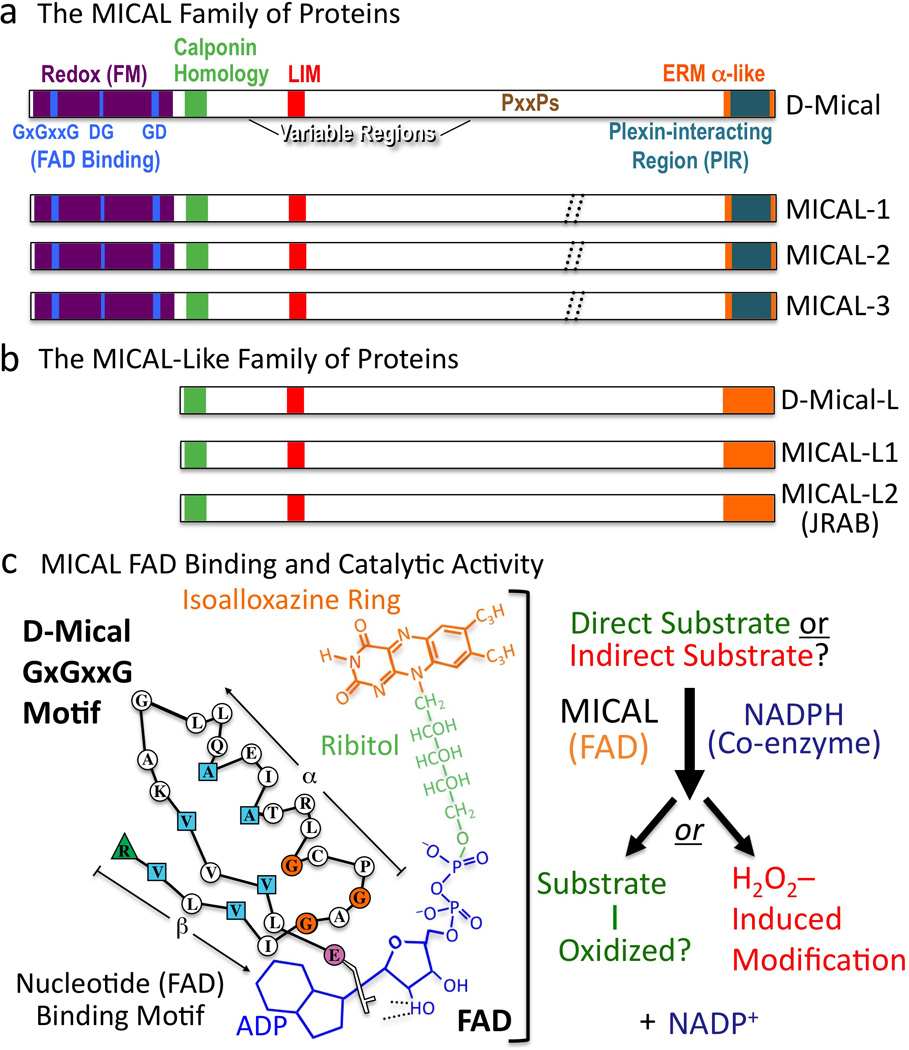
(a) MICAL family proteins are characterized by their flavoprotein monooxygenase (Redox [FM]), calponin homology, LIM, and ERM alpha-like domains. Variable regions differ in length and continuity among family members (dotted //; Box 3). To conform with Drosophila nomenclature guidelines, lowercase lettering is now used when describing invertebrate MICALs (Mical), and all capitals are used when describing the vertebrate MICALs (and the MICAL family of proteins). (b) MICAL-Like proteins are similar to MICALs in domain organization except they lack the Redox domain. (c) FAD binding and MICAL enzymatic activity. (Left) The MICAL cofactor FAD is composed of ADP (including a pyrophosphate group), ribitol, and an isoalloxazine ring. To bind to the ADP region of FAD, Mical relies on its GxGxxG region where the critical residues are colored (Modified from (Wierenga et al. 1986)). To bind to the pyrophosphate and ribitol moieties of FAD, Mical relies on its DG and GD regions, respectively. (Right) Results indicate that the Redox domain of MICAL binds FAD, consumes NADPH, and generates H2O2. MICAL substrate/s are unknown. Alternatively, MICAL may have no direct substrate and may simply generate H2O2.
