Abstract
Background
Semantic interoperability between routine healthcare and clinical research is an unsolved issue, as information systems in the healthcare domain still use proprietary and site-specific data models. However, information exchange and data harmonization are essential for physicians and scientists if they want to collect and analyze data from different hospitals in order to build up registries and perform multicenter clinical trials. Consequently, there is a need for a standardized metadata exchange based on common data models. Currently this is mainly done by informatics experts instead of medical experts.
Objectives
We propose to enable physicians to exchange, rate, comment and discuss their own medical data models in a collaborative web-based repository of medical forms in a standardized format.
Methods
Based on a comprehensive requirement analysis, a web-based portal for medical data models was specified. In this context, a data model is the technical specification (attributes, data types, value lists) of a medical form without any layout information. The CDISC Operational Data Model (ODM) was chosen as the appropriate format for the standardized representation of data models. The system was implemented with Ruby on Rails and applies web 2.0 technologies to provide a community based solution. Forms from different source systems – both routine care and clinical research – were converted into ODM format and uploaded into the portal.
Results
A portal for medical data models based on ODM-files was implemented (http://www.medical-data-models.org). Physicians are able to upload, comment, rate and download medical data models. More than 250 forms with approximately 8000 items are provided in different views (overview and detailed presentation) and in multiple languages. For instance, the portal contains forms from clinical and research information systems.
Conclusion
The portal provides a system-independent repository for multilingual data models in ODM format which can be used by physicians. It serves as a platform for discussion and enables the exchange of multilingual medical data models in a standardized way.
Keywords: Semantic interoperability, CDISC ODM, form exchange, form repository
1. Introduction
Semantic interoperability between routine healthcare and clinical research is a well-known and unsolved issue in the healthcare sector. With growing interdisciplinary collaboration, information exchange becomes even more important. This includes the cooperation between different hospitals, working together in multi-center clinical trials or registries, as well as the cooperation between different departments of the same hospital. The missing interoperability results in redundant documentation and high workloads in times of resource shortage [1]. Despite existing communication standards in healthcare, like those provided by HL7 [2] or Clinical Data Interchange Standards Consortium (CDISC) [3], data exchange typically occurs between information systems of the same institution or hospital information systems (HIS) of the same vendor. In some cases, it is not possible to “exchange CRFs (Case Report Forms) even between different versions of the same software” [4]. This becomes more important as the consolidation of CRFs is hampered and results in disproportional costs.
To exchange metadata from CRF or HIS forms, it is necessary to exchange the respective data models beforehand. We consider a data model to be the technical specification of a medical form, which contains all attributes with respective data types and value lists but without any layout information. In the following, a data model is defined as a common format for data elements [5] and not as a database model. A (medical) form therefore consists of a data model, which is often proprietary, and additional layout information.
2. Objectives
We propose to enable form based metadata exchange. From a physician’s perspective routine and trial documentation consist of a set of items, which are collected in a form. The coding of individual items, as well as the information on which items are selected for in a form, is of importance.
A portal for medical forms may track this issue by allowing for the exchange of data models. Such a portal should be built upon a physician-centered approach, enabling physicians to discuss medical forms and data items needed for their interdisciplinary collaboration. The technical representation of these forms should be based on existing standards.
Therefore, we intend to provide a platform which allows a community-based consensus process on medical data models. Our objective is to implement a web-based portal that
-
•
provides a repository for different types of medical forms (EHR forms, CRFs) in a standard format,
-
•
enables viewing, discussing, rating and exporting of medical forms by healthcare professionals and
-
•
supports the comparison between medical data models.
3. Background
3.1 Current Situation
There are hundreds of HIS vendors worldwide [6] with proprietary data models. Local HIS, initially implemented for billing purposes, are evolving into trans-sectorial Electronic Health Record (EHR) systems with a growing demand to exchange data between different healthcare providers. Typically, most forms of these information systems are customized by form designers resulting in system-specific, site-specific and thus proprietary data items. These data items are often similar as they all cover important aspects of medical documentation, but still differ in their specification (item names, data types) and especially in their coding. However, this coding is the most important aspect to achieve semantic interoperability [7].
Currently, data is mainly stored in dual-source information systems, in which routine documentation is separated from research databases. Missing interoperability between these systems causes additional efforts and incompatibilities in data analysis. This becomes more evident in university hospitals where physicians spend a lot of time on research. From a physician’s point of view, it is especially difficult to understand that so many different documentation systems are available with different forms, even though the medical processes concerning diagnosis, therapy and follow-up documentation are quite similar or even identical. To overcome the problem of separate databases, more and more projects focus on EHR reuse [8] and implementing single source information systems [9–14], in which data items of the routine medical documentation are used for research questions and quality management.
3.2 Prior Work
However, even single source information systems are not able to overcome the interoperability issues between different sites. There are different approaches to tackle this problem which may be grouped regarding their granularity. The following list provides a simplified overview of different options without claiming to be complete.
-
•
Item-based approach:
-
•
Archetype-based approach:
-
•
Information model-based approach:
-
–
For example, the reference information model (RIM) from HL7 [19]
-
–
Up until now the RIM has had major obstacles [20], and many other projects in this area are based on only a few items defined in a metadata repository or a data dictionary. The usage of these metadata repositories is still limited because – amongst others – in a clinical setting, not individual items, but a large set of items are needed. Physicians usually think and work in patient-related documentation tasks and not in single data items. This becomes evident when discussing data item collections for registries that are divided into task specific forms (e.g. informed consent, medical history data). Such a systematic collection of data items being unified in one structure is the main idea of the archetype approach. An archetype intends to be a complete representation of a whole concept which can be instantiated in the form of templates.
3.3 Document Standards
In order to provide a standardized representation, we analyzed document standards within the healthcare domain to define forms with respective attributes and value lists. Therefore, HL7 Clinical Document Architecture (CDA) [21] and CDISC Operational Data Model (ODM) [22] are discussed. Both support multilingual data items, groups of data elements and integration of code systems.
3.3.1 CDA
CDA is a HL7 standard to describe the structure and semantics of clinical documents, with the aim to produce both machine-readable and human-readable documents using XML, HL7 RIM and coded vocabularies. In particular, the specification of the body of CDA Release 2 (R2) documents facilitates semantic interoperability [2]. In addition, CDA R2 offers a simple wrapping mechanism to include a non-XML document within the CDA document [23]. CDA is expected to play an important role for the realization of semantic interoperability. Above all, the ability to share and exchange data collection forms will be very useful “in allowing hospital A to take advantage of a data capture solution designed in hospital B” [24].
3.3.2 ODM
ODM is a well-established, XML-based CDISC-standard, which is commonly used in clinical trials for example, to archive data and metadata of clinical trials [22, 29]. One of the CDISC core principles is to “work with other professional groups to encourage that there is maximum sharing of information“ [3]. Their mission is “to develop and support global, platform-independent data standards that enable information system interoperability to improve medical research and related areas of healthcare“ [3]. The ODM-format is system independent and vendor neutral which is important for form discussion and form exchange. In addition, CDISC standards are postulated by the government (e.g. Food and Drug Administration (FDA) trial submission) and ODM is used in different electronic data capture (EDC) systems, as well as in clinical data management systems (CDMS) used by the pharmaceutical industry (e.g. Rave [25], OpenClinica [26], Marvin [27]). Especially in the context of clinical trials, CDISC standards are more established and many EDC systems already support ODM files [28].
3.4 Coding Standards
3.4.1 Terminologies
In addition to the syntactical specification, the attributes of our medical data models need an underlying data item definition. We used the terminology codes from Systematized Nomenclature in Medicine – Clinical Terms (SNOMED CT) [30], Unified Medical Language System (UMLS) [31] and Logical Observation Identifiers Names and Codes (LOINC) [32] to assure semantic interoperability on the level of data elements. To enable semantic interoperability between different systems, standardized “coding” is advantageous. However, the term “coding” has different meanings. On the one hand, it is used to specify value sets for dedicated data items, such as International Classification of Diseases (ICD) codes for diagnoses [33, 34], or the Medical Dictionary for Regulatory Activities (MedDRA) for adverse events [35]. On the other hand, “coding” can refer to code items themselves. In this case, codes represent a medical concept and facilitate that two sites of a clinical trial collect the same data item. Terminology systems like SNOMED CT and LOINC seem to be more adequate for detailed medical documentation and laboratory codes.
3.4.2 CDASH
With the Clinical Data Acquisition Standards Harmonization (CDASH) project there exists an approach to describe basic standards for the collection of clinical trial data [36]. CDASH is based on the CDISC Study Data Tabulation Model (SDTM) [37] and describes fields for 18 domains of medical research (e.g. demographics, adverse events). CDASH should establish a standardized data collection baseline for clinical trial submissions.
3.4.3 BRIDG
Another approach of harmonizing existing standards is proposed by the Biomedical Research Integrated Domain Group (BRIDG). To support interoperability between research and healthcare they developed an overarching model which contains streams of development of CDISC, NCI, HL7 and the FDA [38, 39].
Our analyses showed that both CDA and ODM provide suitable representations for a web-based form repository. Therefore, the decision was taken to initially use the CDISC ODM format (ODM v1.3.1) as a main document standard for our portal, and in a second step, to integrate CDA by using the BRIDG model to harmonize data elements. The interoperability between CDA and ODM was already shown by El Fadly et al in 2007 [40].
4. Methods
Basic requirements for the implementation of the web-based portal were collected in non-structured and informal interviews with physicians and medical informatics experts. In addition, existing
EDC systems were taken into account to specify the portal for medical data models. Use-cases were identified and modeled in UML. The internal data structure is represented in an entity-relationship diagram (ER-diagram). Based on this information the web-based portal was designed as follows.
4.1 Implementation
Ruby on Rails (Ruby version 1.9.2, Rails version 3.1) [41] was chosen to implement a modern web 2.0 platform. Rails is an open source framework with an intuitive programming style that allows for rapid software prototyping. The layout of a Rails application strictly follows the MVC pattern (model/view/controller). Ruby on Rails is platform independent, runs on any server operating system and there are several ways of application scaling.
4.2 Usability Criteria
The software system sticks to the usability criteria defined in DIN EN ISO 9241. Part 11 of this standard defines three guidance criteria (effectiveness, efficiency and the user’s satisfaction) that were followed during the conception, planning and design of the system. As the satisfaction of a user has a subjective component, part 12 of the standard that defines rules for the presentation of information (clarity, discriminability, compactness, consistency, perceptibility, legibility and perceivability) was also taken into account [42].
4.3 Form Conversion
In particular, EHR forms of the local hospital and from the open source HIS OpenVista [43] were converted into ODM format. Forms of the local hospital were manually mapped to ODM. The conversion of OpenVista forms to ODM was done with a dedicated converter and manual curation. A further conversion was performed by transforming CRFs from NCIs (National Cancer Institute) Cancer Data Standards Registry and Repository into CDISC ODM, reaching interoperability in clinical research [44]. A subset of converted CRFs was uploaded into the portal.
In addition, inclusion and exclusion criteria from local studies available from the NIH clinical trials registry [45] were manually transformed by physicians and linguists into ODM and uploaded into the portal. An automated transformation was not possible as eligibility information was only available as free text.
5. Results
The main focus of the form-based approach is the integration of the physician. The following list shows important implementation aspects which were summarized after informal interviews with physicians and expert-discussions with health informatics professionals.
5.1 Implementation Aspects
Functional aspects
-
•
Form based approach: The structure of the portal has to follow a form-based approach and not focus on single data elements because physicians are used to working with forms.
-
•
Compatible with EDC systems: To support clinical research it is important that the portal is compatible with common EDC-systems.
-
•
Applicable to different types of forms: It should be possible to upload different types of forms so that the portal may contain CRFs as well as EHR forms, because users want to discuss their data elements independently from their information systems.
-
•
Version Control: Forms are regularly updated, which results in many versions of the same form. Changes should be tracked in a history.
-
•
Multiple languages: The portal is designed for a wide range of users so it should be possible to switch between common languages. At the beginning, at least English and German have to be supported.
Non-functional aspects
-
•
Usability: The portal has to follow common usability criteria like learnability, efficiency and satisfaction so that it can be easily used by physicians.
-
•
Community based: User content is very important. Physicians and other health care professionals should be able to upload their forms for discussion.
-
•
System independent: Access to the portal should be independent from the operating system, the browser and security settings (e.g. in the hospital).
5.2 Use Cases
Based on the physician interviews three main use cases for the portal were identified:
-
•
Upload a form: The physician has to be able to provide his own data models for discussion.
-
•
View form: The physician has to view published forms of other users in an overview and in a detailed view which should also include:
-
–
Search: The physician has to be able to search for topics and forms.
-
–
Comment: The physician has to be able to comment other forms.
-
–
-
•
Download form: The physician has to be able to download forms (as a PDF, CSV or ODM file).
5.3 Data Structure
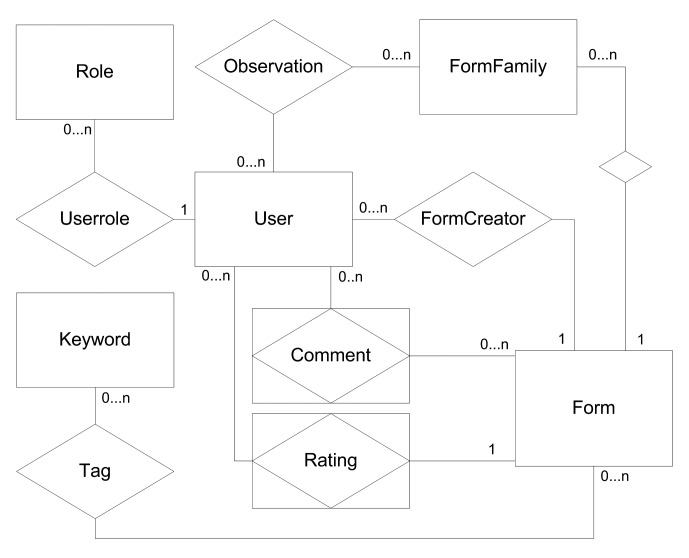
Based on the requirements and use-cases, a data base for the portal was developed. The main part is illustrated in an entity-relationship model (Fig. 1). The key element of the physician-centered approach is the user who can create, observe and make comments on the forms. Each form belongs to a form family to support versioning. In addition, each form can be tagged with keywords to allow a more specific search.
Fig. 1.
ER-diagram of the web-based application. Forms belong to form families, have one creator (owner) and many observers and may be tagged with different keywords. Users belong to roles to differentiate moderators and reading users.
5.4 Portal for Medical Data Models
Based on these classes, data structures, use cases and implementation aspects, a web-based portal for medical data models was implemented and can be accessed under the following URL: http://www.medical-data-models.org
Two physicians already uploaded forms for testing purposes. Currently, forms of the local HIS and from the open source HIS OpenVista are included. In addition, EDC forms, a subset of converted NCI forms and eligibility forms, are available. Overall, more than 250 forms with approximately 8000 items are provided by the system. Some examples are presented in the following list:
-
•
Braden Scale (converted form from the open source HIS OpenVista)
-
•
Cardiovascular medical history (form from the local HIS)
-
•
Follow-up form (research form from the local HIS)
-
•
Eligibility NCT00006045 AML (Eligibility criteria form originated from ClinicalTrials.gov)
Users are able to browse and view forms. Registered users are able to comment and rate forms. ►Figure 2 shows the overview of the portal. On the left side basic descriptive statistics about the content are presented, particularly the total number of available data models, item groups and items. The main area contains the list of available forms, which are presented with their name, version (in brackets), date of upload, author and latest comments. The search function allows a quick finding of the relevant forms.
Fig. 2.
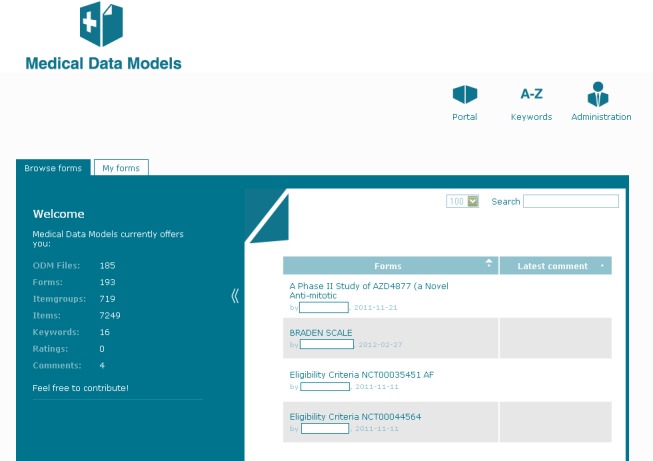
Overview of medical data forms in the portal. Basic statistic information is presented on the left side. All forms are presented with author, upload date and latest comment.
Each form is presented in a form-data view (only attributes) and in a detailed view (attributes + data types + value sets). The form-data view (►Figure 3) shows all items, separated by item groups and respective input fields. Required input fields are marked with an asterisk. On the left side, metadata of the form are presented. These include information about title, author and keywords. Furthermore, the user is able to update a form, switch the language of the form content, or download the form in three different formats (as PDF, CSV or ODM file). In addition, it is possible to leave a comment.
Fig. 3.

Form data view. On the left side, metadata like author and keywords are presented. The main screen shows all items with respective input fields grouped by the ODM item groups. Corresponding to the data types the respective input fields are displayed.
►Figure 4 presents a form in a detailed view. In addition to the items and groups from the form-data view, data type and coding are also displayed. The coding shows the aliases and code systems which are specified in the ODM file. In this example it contains SNOMED CT codes, UMLS identifier, LOINC and MedDRA codes.
Fig. 4.
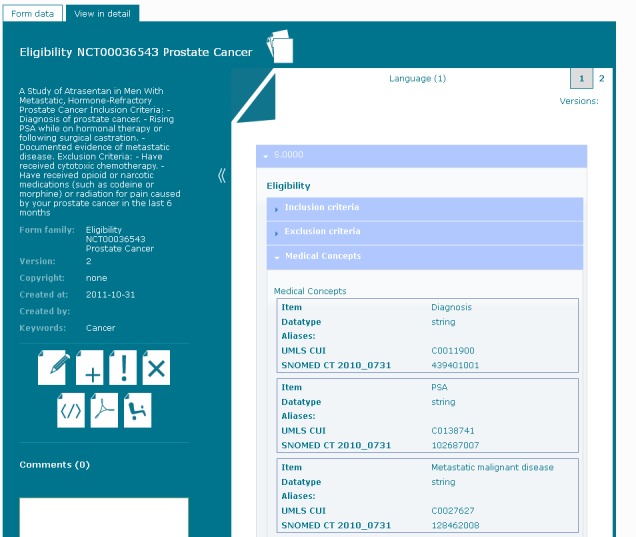
Form view in detail. The detailed view displays all items grouped by the specified item groups. In addition all items are presented with data type and item codes.
6. Discussion
The implemented web-based portal presents a form-based approach to store and discuss medical data models. Initial testing demonstrates that this approach is suitable for different types of forms (EHR forms, eligibility forms and research forms). Physicians are now able to upload, comment and rate item sets in order to support semantic interoperability, which is required to enhance the collaboration between clinical partners.
A full-scale HIS contains a huge number of items which are subject to change over time. The discussion and harmonization of different languages and code systems results in an effort that cannot be managed by a single person or group. Therefore, the publication and international discussion of data items or data models should be community driven. The portal applies the concept of the web 2.0, especially the aspect of user driven content. Therefore, the benefits of this portal are expected to grow with the number of users and uploaded content, just as Wikipedia grows with the number of involved authors and articles.
6.1 Related work
Pathak et al. demonstrate the need for semantic interoperability, but focus more on the coding of single items [46]. Another interesting method is the use of a hierarchic attribute-value scheme for metadata exchange, as described by Gardner et al. [47]. They want to enable interoperability at different depths of their variable tree, but their approach is a proprietary solution. On the contrary, Carlson et al. focus on healthcare standards, and use HL7 CDA to derive a specific model for information exchange [48]. Their approach is similar to our solution as they also place emphasis on closing the gap between the clinical domain experts and the healthcare IT domain experts. Kunz et al. work on the interoperability across biomedical databases by using a repository of Common Data Elements (CDEs) contributes to the implementation of interoperable systems [49]. This approach can succeed if the amount of available data elements is high enough. El Fadly et al. also address interoperability issues, but focus more on a direct conversion of data, while our objective was to build a repository of forms in a standard format [40].
6.2 Strengths and Limitations
From our perspective, many existing interoperability concepts like the archetype approach, the item-based metadata repository and the HL7 RIM [15–19] are too complicated for healthcare professionals and therefore rarely used in clinical context. Input from healthcare professionals is mandatory to define medical data models. Therefore, it is required to foster exchange and enable consensus discussions, preferably on an international level with multilingual support. The portal for medical data models provides a platform which allows for user driven form-exchange and discussion. The use of established standards like ODM will increase acceptance because it is a well-known standard used in several commercial CDMS or EDC systems. In addition, the user interface was designed to enable form upload with a few simple steps. An important aim of this portal is the support of clinical trials, for instance, the management of inclusion and exclusion criteria. Eligibility forms can be uploaded into the portal in ODM format, compared with other available eligibility forms and commented by the users.
So far the developed portal is only a prototype to present the idea of the form based approach. Although it contains a lot of forms which were uploaded mainly by local physicians, it has not yet been formally evaluated by the target user group (physicians), or in its real clinical context. As ODM is currently not provided as metadata standard in most EHR systems, there is still the issue that forms need to be manually extracted and converted into ODM format before they can be uploaded into the repository. Therefore, tools to support handling of ODM files for physicians are needed.
6.3 Future Research
To analyze the clinical acceptance of the system, an evaluation study for a broader user community is planned with a special focus on usability aspects.
A key issue of future research is the integration of ODM-supporting components, for example a tool to enable coding of data items within an ODM file. Another implementation aspect is the compatibility with statistics software. Once it is possible to extract information about variables and data types from this platform, ODM files could be used to create corresponding database schemes. For sustainability, the portal needs to be adapted to upcoming ODM versions. Currently, forms in ODM format have to be manually built with text- or XML-editors. An ODM editor or a tool for converting different formats into ODM would be helpful to process forms. Conversion tools between ODM and CDA form representations are another important topic for future research to facilitate the exchange of forms with various EHR systems. In addition to the document structure, it is also important to support coding of the items and value lists. There already exists terminology services like Lex-EVS [46, 50] and NCBO Bioportal [51, 52], with respective APIs which can be used to support the coding.
7. Conclusion
A form-based approach for metadata exchange via data models of EHR forms and CRFs in the same system is technically feasible. Consequently, a system-independent platform to store and share multilingual data models based on international standards can be provided for physicians.
Clinical Relevance Statement
A repository of open medical data models based on existing standards in healthcare, such as ODM, can provide a platform to exchange and discuss clinical as well as research forms. If adopted by the medical community and software providers, this user-driven web 2.0 approach can contribute to semantic interoperability and thus collaboration of physicians across different sites.
Conflict of Interest
There are no conflicts reported by any of the authors.
Human Subjects Protections
We acknowledge that neither human nor animal subjects were included in the project.
Acknowledgments
Javier García supported the design of the web-portal. Olivia Lee supported English Writing.
References
- 1.Dugas M, Roeder N.Entlastung des medizinischen Personals bei der Dokumentationsarbeit im Krankenhaus. Das Krankenhaus 2011; 103(1): 35- [Google Scholar]
- 2.Health Level Seven (HL7) [Internet], [cited 2012. Mar 22]. Available from: http://www.hl7.org
- 3.Clinical Data Interchange Consortium (CDISC) [Internet], [cited2012Mar 22]. Available from: http://www.cdisc.org
- 4.Kuchinke W, Wiegelmann S, Verplancke P, Ohmann C. Extended cooperation in clinical studies through exchange of CDISC metadata between different study software solutions. Methods Inf Med 2006; 45(4): 441–446 [PubMed] [Google Scholar]
- 5.Reisinger SJ, Ryan PB, O'Hara DJ, Powell GE, Painter JL, Pattishall EN, et al. Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases. J Am Med Inform Assoc 2010; 17(6): 652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eurorec [Internet], [cited2012Mar 22]. Available from: http://www.eurorec.org/
- 7.Oemig F, Blobel B. Semantic interoperability adheres to proper models and code systems. A detailed examination of different approaches for score systems. Methods Inf Med 2010; 49(2): 148–155 [DOI] [PubMed] [Google Scholar]
- 8.Prokosch HU, Ganslandt T.Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med 2009; 48(1): 38–44 [PubMed] [Google Scholar]
- 9.Kush R, Alschuler L, Ruggeri R, Cassells S, et al. Implementing Single Source: the STARBRITE proof-of-concept study. J Am Med Inform Assoc 2007, 14(5): 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breil B, Semjonow A, Müller-Tidow C, Fritz F, Dugas M.HIS-based Kaplan-Meier plots – a single source approach for documenting and reusing routine survival information. BMC Med Inform Decis Mak 2011; 11: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Fadly A, Lucas N, Rance B, Verplancke P, Lastic PY, Daniel C. The REUSE project: EHR as single data-source for biomedical research. Stud Health Technol Inform. 2010; 160(Pt 2): 1324–1328 [PubMed] [Google Scholar]
- 12.Murphy EC, Ferris FL, 3rd, O'Donnell WR. An electronic medical records system for clinical research and the EMR EDC interface. Invest Ophthalmol Vis Sci 2007; 48(10): 4383–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritz F, Ständer S, Breil B, Riek M, Dugas M. CIS-based registration of quality of life in a single source approach. BMC Med Inform Decis Mak 2011; 11: 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziuballe P, Forster C, Breil B, Thiemann V, et al. The single source architecture x4T to connect medical documentation and clinical research. Stud Health Technol Inform 2011; 169: 902–906 [PubMed] [Google Scholar]
- 15.Stausberg J, Löbe M, Verplancke P, Drepper J, et al. Foundations of a metadata repository for databases of registers and trials. Stud Health Technol Inform 2009; 150: 409–413 [PubMed] [Google Scholar]
- 16.Jiang G, Solbrig HR, Iberson-Hurst D, Kush RD, Chute CG. A collaborative framework for representation and harmonization of clinical study data elements using semantic MediaWiki. AMIA Summits Transl Sci Proc 2010; 2010: 11–15 [PMC free article] [PubMed] [Google Scholar]
- 17.Kohl CD, Garde S, Knaup P. Facilitating secondary use of medical data by using openEHR archetypes. Stud Health Technol Inform 2010; 160(Pt 2): 1117–1121 [PubMed] [Google Scholar]
- 18.Martínez-Costa C, Menárguez-Tortosa M, Fernández-Breis JT. An approach for the semantic interoperability of ISO EN 13606 and OpenEHR archetypes. J Biomed Inform 2010; 43(5): 736–746 [DOI] [PubMed] [Google Scholar]
- 19.Jenders RA, Sujansky W, Broverman CA, Chadwick M. Towards improved knowledge sharing: assessment of the HL7 Reference Information Model to support medical logic module queries. Proc AMIA Annu Fall Symp 1997: 308–312 [PMC free article] [PubMed] [Google Scholar]
- 20.Smith B, Ceusters W. HL7 RIM: an incoherent standard. Stud Health Technol Inform 2006; 124:133–138 [PubMed] [Google Scholar]
- 21.CDA [Internet], [cited2012Mar 22]. Available from: http://www.cdisc.org/odm
- 22.ODM [Internet], [cited2012Mar 22]. Available from: http://www.cdisc.org/odm
- 23.Dolin RH, Alschuler L, Boyer S, Beebe C, et al. HL7 Clinical Document Architecture, Release 2. J Am Med Inform Assoc 2006; 13(1): 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Fadly A, Rance B, Lucas N, et al. Integrating clinical research with the Healthcare Enterprise: from the RE-USE project to the EHR4CR platform. J Biomed Inform 2011; 44 (Suppl. 1): S94–S102 [DOI] [PubMed] [Google Scholar]
- 25.Medidata Rave [Internet], [cited2012Mar 22]. Available from: http://www.mdsol.com/products/rave_overview.htm
- 26.OpenClinica [Internet], [cited2012Mar 22]. Available from: https://community.openclinica.com/
- 27.Xclinical [Internet], [cited2012Mar 22]. Available from: www.xclinical.com/
- 28.XML 4 Pharma [Internet], [cited2012Mar 22]. Available from: http://www.xml4pharma.com/ODMinEDC/Samples.html
- 29.Kuchinke W, Aerts J, Semler SC, Ohmann C. CDISC standard-based electronic archiving of clinical trials. Methods Inf Med 2009; 48(5): 408–413 [DOI] [PubMed] [Google Scholar]
- 30.SNOMED CT [Internet], [cited2012Mar 22]. Available from: http://www.ihtsdo.org/
- 31.UMLS [Internet], [cited2012Mar 22]. Available from: http://www.nlm.nih.gov/research/umls
- 32.LOINC [Internet], [cited2012Mar 22]. Available from: http://loinc.org/
- 33.DIMDI [Internet], [cited2012Mar 22]. Available from: http://www.dimdi.de
- 34.WHO Classifications [Internet], [cited2012Mar 22]. Available from: http://www.who.int/classifications
- 35.MedDRA [Internet], [cited2012Mar 22]. Available from: http://meddraso.com
- 36.CDASH [Internet], [cited2012Mar 22]. Available from: http://www.cdisc.org/cdash
- 37.SDTM [Internet], [cited2012Mar 22]. Available from: http://www.cdisc.org/sdtm
- 38.BRIDG [Internet], [cited2012Mar 22]. Available from: http://www.bridgmodel.org/
- 39.BRIDG [Internet], [cited2012May 15]. Available from: http://www.cdisc.org/bridg
- 40.El Fadly A, Daniel C, Bousquet C, Dart T, et al. Electronic healthcare record and clinical research in cardiovascular radiology. HL7 CDA and CDISC ODM interoperability AMIA Annu Symp Proc 2007; Oct 11: 216–220 [PMC free article] [PubMed] [Google Scholar]
- 41.Ruby on Rails [Internet], [cited2012Mar 22]. Available from: http://rubyonrails.org/
- 42.INTERNATIONAL ISO STANDARD 9241–11, [Internet], [cited2012Mar 22]. Available from: http://www.it.uu.se/edu/course/homepage/acsd/vt09/ISO9241part11.pdf
- 43.OpenVista [Internet], [cited2012Mar 22]. Available from: http://sourceforge.net/projects/openvista/
- 44.Bruland P, Breil B, Fritz F, Dugas M. Interoperability in clinical research: From metadata registries to semantically annotated CDISC ODM. Accepted for oral presentation at “MIE 2012” [PubMed] [Google Scholar]
- 45.Clinical Trials [Internet], [cited2012Mar 22]. Available from: http://www.clinicaltrials.gov/
- 46.Pathak J, Wang J, Kashyap S, Basford M, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc 2011; 18(4): 376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner D, Knuth KH, Abato M, Erde SM, et al. Common data model for neuroscience data and data model exchange. J Am Med Inform Assoc 2001; 8(1): 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlson D, Farkash A, Timm JT. A model-driven approach for biomedical data integration. Stud Health Technol Inform 2010; 160(Pt 2): 1164–1168 [PubMed] [Google Scholar]
- 49.Kunz I, Lin MC, Frey L. Metadata mapping and reuse in caBIG. BMC Bioinformatics 2009; 10 (Suppl. 2): S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LexEVS Server and API, National Cancer Institute [Internet], [cited2012Mar 24]. Available from: https://cabig.nci.nih.gov/community/tools/LexEVS_Server
- 51.Musen MA, Noy NF, Shah NH, Whetzel PL, et al. The National Center for Biomedical Ontology. J Am Med Inform Assoc 2012; 19(2): 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.NCBO Bioportal. [Internet], [cited2012Mar 24]. Available from: http://bioportal.bioontology.org/